
 |
ChromothripsisDB: a curated database of chromothripsis | ||||||||
| Home | Search | Browse | Download | Tutorial | Contact | ||||
| Tutorial | ||||
1. Browse Studies Related to Chromothripsis 2. Detect Chromothripsis in a Disease of Interest 3. Determine Chromothripsis Data that Is Generated by a Certain Technology 4. Detect Chromothripsis Pattern in Glioma |
||||
| 1. Browse Studies Related to Chromothripsis | ||||
|
A. Click on the Browse menu bar to invoke the Browse page. B. Select “Article Type”. All articles were classified into 3 types: 1) Research. Publications report chromothripsis cases; 2) Review and Opinion. Reviews about chromothripsis in general or chromothripsis in specific disease; 3) Methodology. Publications describe methods or tools to predict or identify chromothripsis phenomenon. C. Main information of each study. PubMed ID was used as the identifier. Click on the study title, the database will return a study information page listing main feature of this study. D. If raw array or sequencing data was submitted to the public databases by the authors, there will be the hyperlinks to related data sources. E. The definition of chromothripsis that is given in the publication. This information was extracted and normalized from the article. The three most significant features shared by different operational chromothripsis definitions were used: 1) the minimum number of close-by breakpoints. 2) the number of copy number states. 3) the random joining of chromosome fragments. F. If copy number aberration and/or breakpoint junction data is provided in the article, there will be the option of downloading data files in text format. G. If the publication is a research article, chromothripsis cases included in this article will be displayed when the data is available. Genomic copy number aberration of each chromosome is shown. H. Circos plot displays the rearrangements in disease samples and links between translocations. |
|||
|
||||
| top | ||||
| 2. Detect Chromothripsis in a Disease of Interest | ||||
A. Click on the Search menu bar to invoke the Search page. B. Select “All” or any specific species in the Species field. Use Command + click (Mac) or Control + click (PC) to select multiple menu options. C. Select “All” or any specific technologies in the Technology field. D. Select any specific diseases in the Disease Type field. Use Command + click (Mac) or Control + click (PC) to select multiple menu options. E. Click on Search. F. The database will return a results page listing every case that is eligible for search parameters. The general information of search results will be shown. If chromothripsis data is available, the results page will include a hyperlink to download the data. G. Statistics figures will show the distribution of shattered chromosomes and percentage of disease types. H. Each Chromothripsis case that is related to selected disease types will be displayed when the data is available. |
||||
|
||||
| top | ||||
| 3. Determine Chromothripsis Data that Is Generated by a Certain Technology | ||||
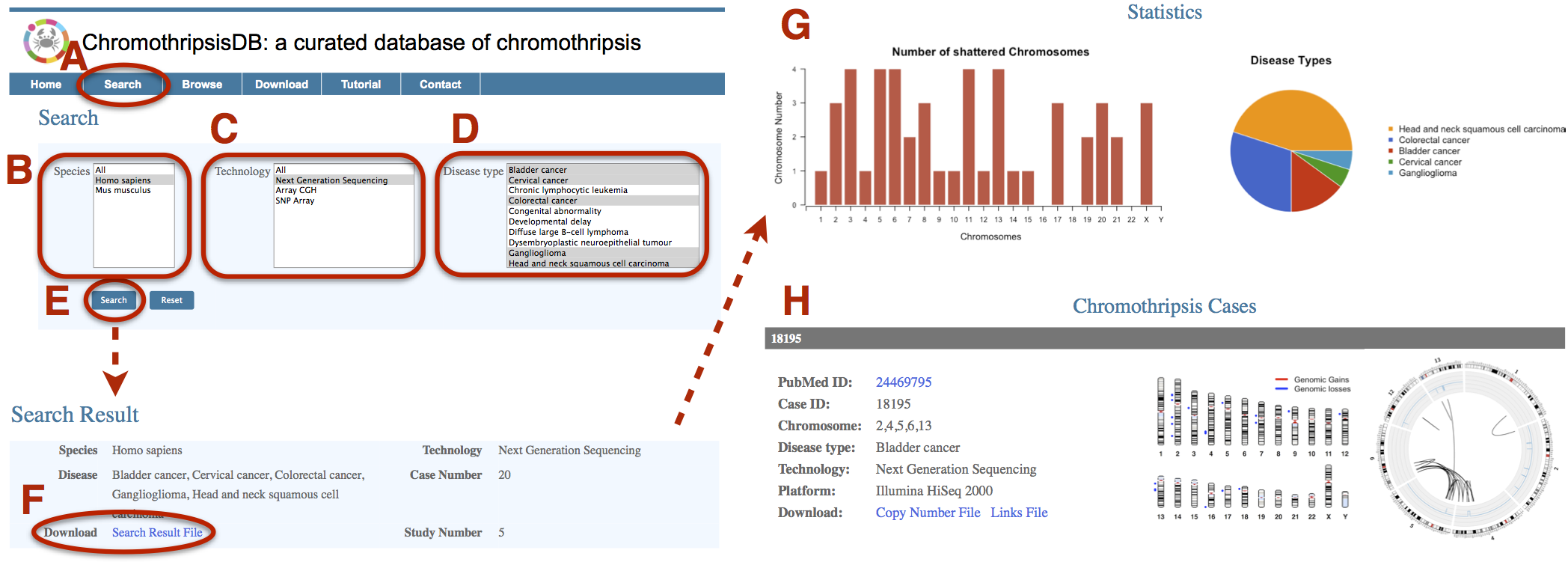
A. Click on the Search menu bar to invoke the Search page. B. Select “All” or any specific species in the Species field. C. Select any specific technologies in the Technology field. D. Select “All” or any specific diseases in the Disease Type field. Use Command + click (Mac) or Control + click (PC) to select multiple menu options. E. Click on Search. F. The database will return a results page listing every case that is eligible for search parameters. The general information of search results will be shown. If chromothripsis data is available, the results page will include a hyperlink to download the data. G. Statistics figures will show the distribution of shattered chromosomes and percentage of disease types. H. Each Chromothripsis case that is generated by selected technologies will be displayed when the data is available. |
||||
 |
||||
| top | ||||
| 4. Detect Chromothripsis Pattern in Glioma | ||||
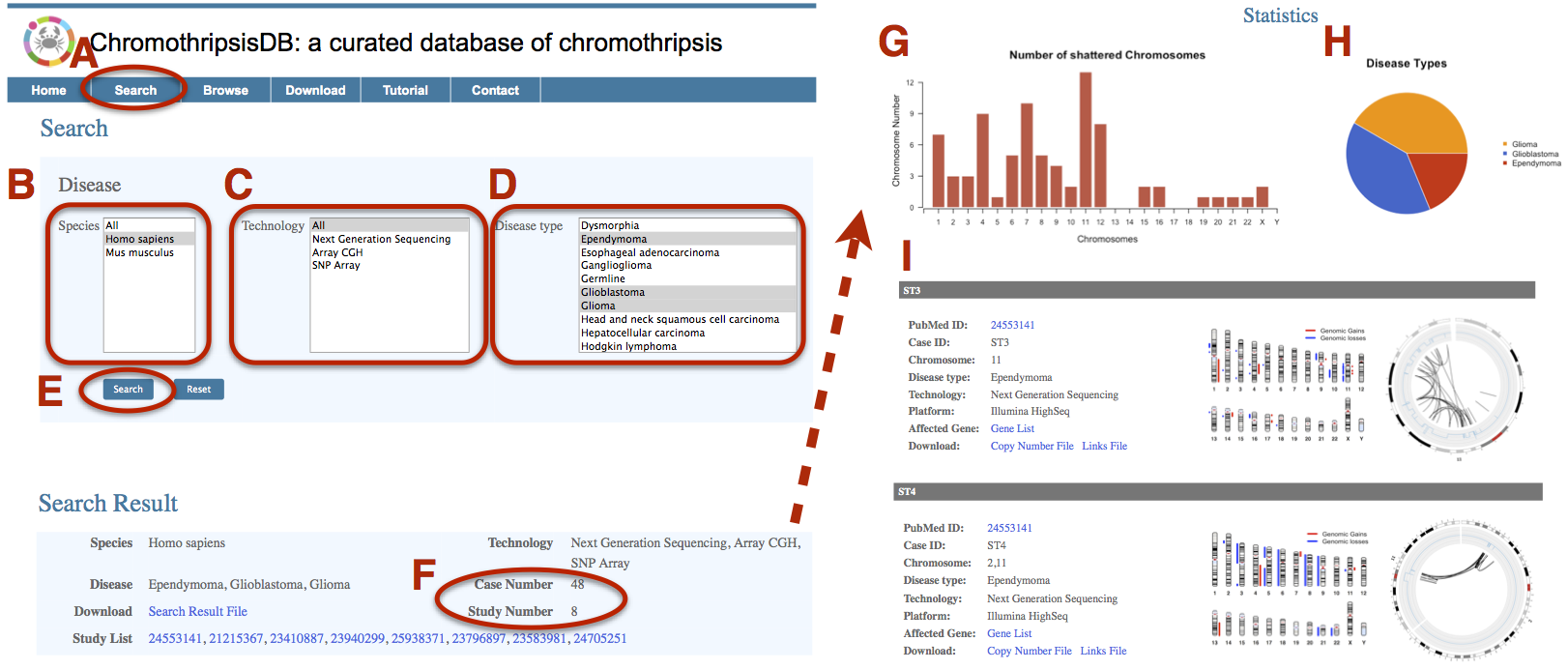
ChromothripsisDB can be used to conveniently obtain disease specific chromothripsis patterns. Glioma is a common type of brain tumor and has three subtypes in our database: Ependymoma, Glioblastoma and Glioma. Here, we detect chromothripsis pattern in gliomas. A. Click on the Search menu bar to invoke the Search page. B. Select “Homo sapiens” in the “Species” field. C. Select “All” in the “Technology” field. D. Select Ependymoma, Glioblastoma and Glioma in the Disease Type field. Use Command + click (Mac) or Control + click (PC) to select multiple menu options. E. Click on Search. F. The result page shows 48 cases collected from 8 publications. G. The bar plot displays the distribution of pulverized chromosomes across the genome. Here, the most commonly shattered chromosome is chromosome 11, which is a prominent hot-spot for chromothripsis in glioma. Other chromosomes with a remarkably high frequency of pulverization include chromosome 4 and 7. H. The pie chart shows the percentage distribution of tumor samples among the three gliomas. I. Related Chromothripsis cases are displayed. The chromosome idiogram and Circos plot intuitively show the copy number aberrations and structural variations. Here we present the sample ST3 that contains a single whole chromosome 11 shattering, and the sample ST4 that has localized genome fragmentation, but involves two chromosomes. |
||||
 |
||||
| top | ||||
| 5. Detect Common Affected Genes in Cancers | ||||
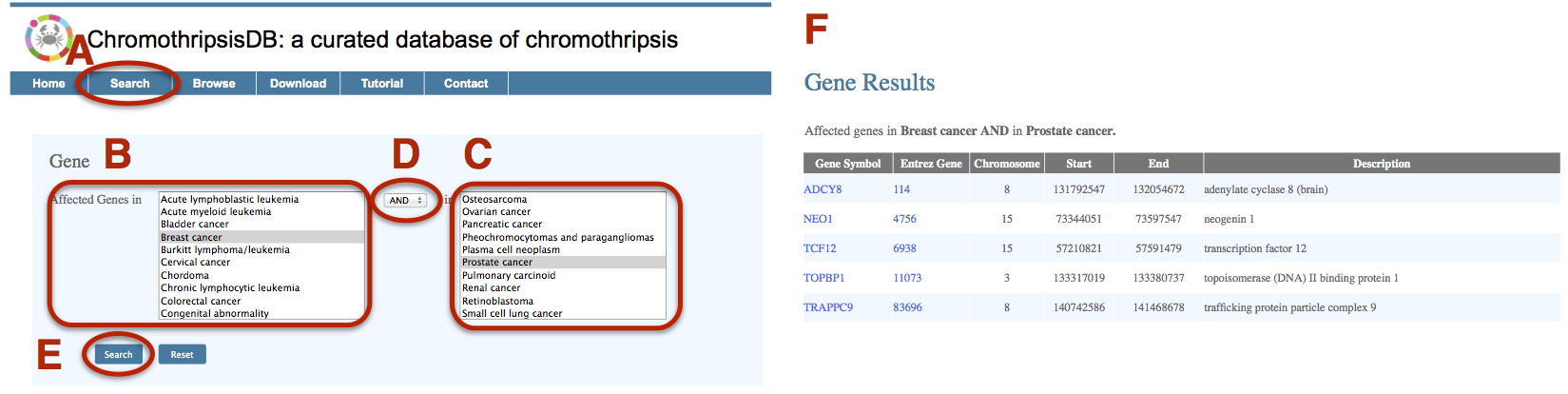
The detection of affected genes in various cancer types will facilitate to narrow the gene list to the most promising ones, which are involved in the chromosome shattering event. Here, we detect the affected genes between breast cancer and prostate cancer. A. Click on the Search menu bar to invoke the “gene search” interface. B. Select “Breast cancer” in the first disease field. C. Select “Prostate cancer” in the second disease field. D. Choose “AND” as the Boolean operator in the drop-down box. E. Click on Search. F. The result page shows a gene list table containing the location and description of each gene, as well as hyperlinks to other resources. |
||||
 |
||||
| top | ||||
| 6. Detect Cancer Type Specific Genes | ||||
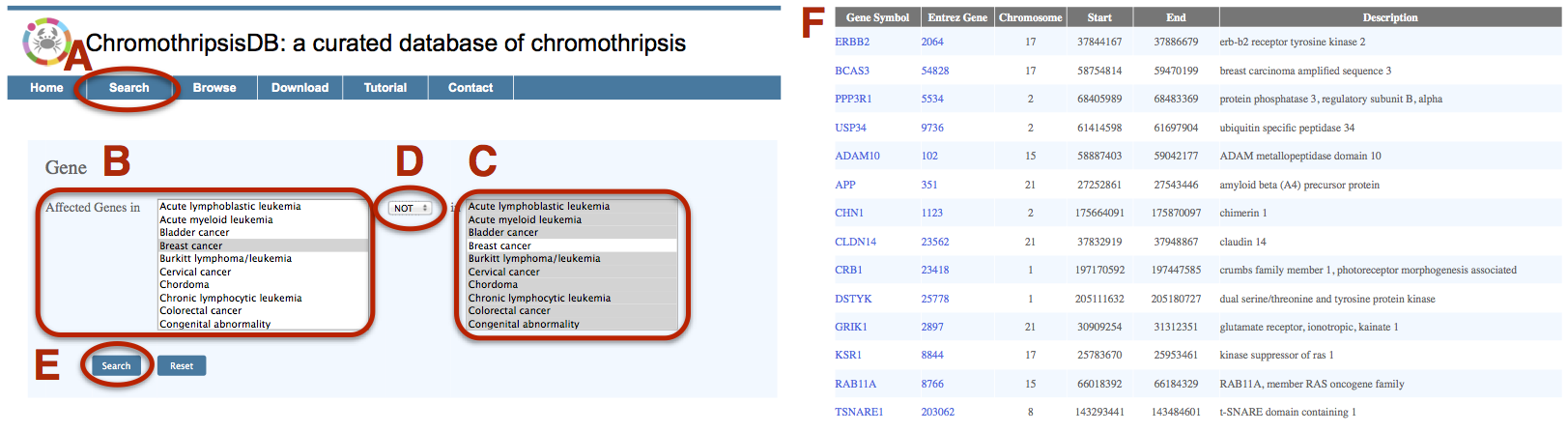
The detection of affected genes in a certain cancer type will facilitate the identification of markers associated with the cancer initiation and development. Here, we queried the breast cancer specific genes from the database. A. Click on the Search menu bar to invoke the “gene search” interface. B. Select “Breast cancer” in the first disease field. C. Select all other cancers in the second disease field. D. Choose “NOT” as the Boolean operator in the drop-down box. E. Click on Search. F. The result page lists breast cancer specific genes that involved in the chromothripsis region. The page also shows the corresponding gene annotations, as well as hyperlinks to other resources. |
||||
 |
||||
| top | ||||
| Chromothripsis database. 2025 Cai Laboratory |