 |
MethCNA: a database for integrating genomic and epigenomic data in cancer | |||||||
| Home | Search | Browse | Cluster | Tutorial | Contact | |||
| MethCNA | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
MethCNA is a comprehensive database for the integrated analysis of DNA methylation and copy number alterations in human cancer. Currently, the database contains about 10,000 publicly available tumor samples interrogated by Illumina Infinium HumanMethylation450 BeadChip. The Infinium HumanMethylation450 BeadChip is based on similar biochemical reaction principle and technology as the single nucleotide polymorphism arrays, and is able to robustly detect CNAs as a no-cost byproduct of methylation studies. The raw array data were collected from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases and preprocessed with processing pipeline. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Genomic & Epigenomic Data Integration | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
MethCNA is a resource for large-scale integration analysis of genomic and epigenomic data in human cancer. The integration of different omics data types is an important approach for understanding the fundamental mechanisms of cancer development. DNA methylation and copy number alteration data from the same tumor specimen may allow the identification of synergistic mechanisms for the inactivation of tumor suppressor genes or the activation of oncogenic pathways. The efficient mining of these datasets may provide valuable insight into the underlying molecular bases of oncogenesis, and facilitate to identify driver from passenger alterations. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
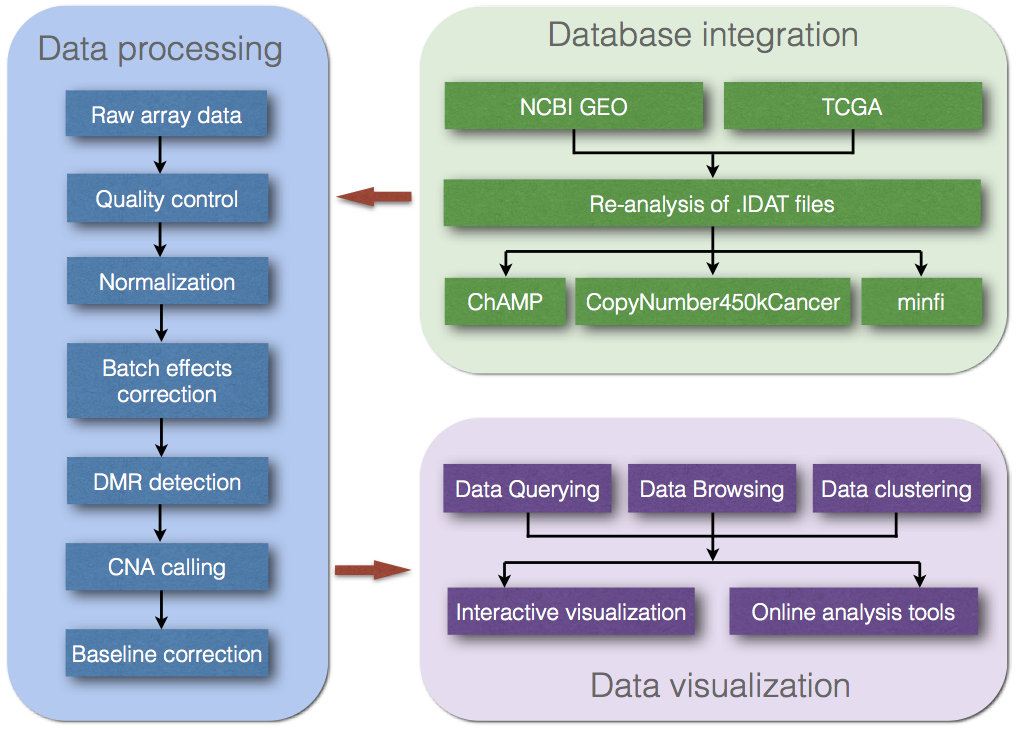
| Data Processing Pipeline | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The raw signal intensity data (.IDAT) files are reanalyzed to obtain normalized data. The data processing pipeline integrates a collection of widely used R packages, including ChAMP, CopyNumber450kCancer and minfi. It comprises several analysis steps, including normalization, quality control, batch effect analysis, copy number aberrations calling, detection of differentially methylated regions, baseline correction and so on. The regions of CNA and the sites of aberrant CpG methylation are deposited in MethCNA and user-interface are developed. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Database Contents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 1. The Illumina Infinium HumanMethylation450 BeadChip array integrated in MethCNA
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| © 2017 Cai Laboratory. Last updated on 30 December 2017 | |||||||||