| Full name: olfactory receptor family 6 subfamily X member 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11q24.1 | ||
| Entrez ID: 390260 | HGNC ID: HGNC:14737 | Ensembl Gene: ENSG00000221931 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
OR6X1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04740 | Olfactory transduction |
Expression of OR6X1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | OR6X1 | 390260 | 78488 | 0.0615 | 0.3745 | |
| GSE53624 | OR6X1 | 390260 | 78488 | 0.1812 | 0.0321 | |
| GSE97050 | OR6X1 | 390260 | A_33_P3384902 | 0.1635 | 0.5268 |
Upregulated datasets: 0; Downregulated datasets: 0.
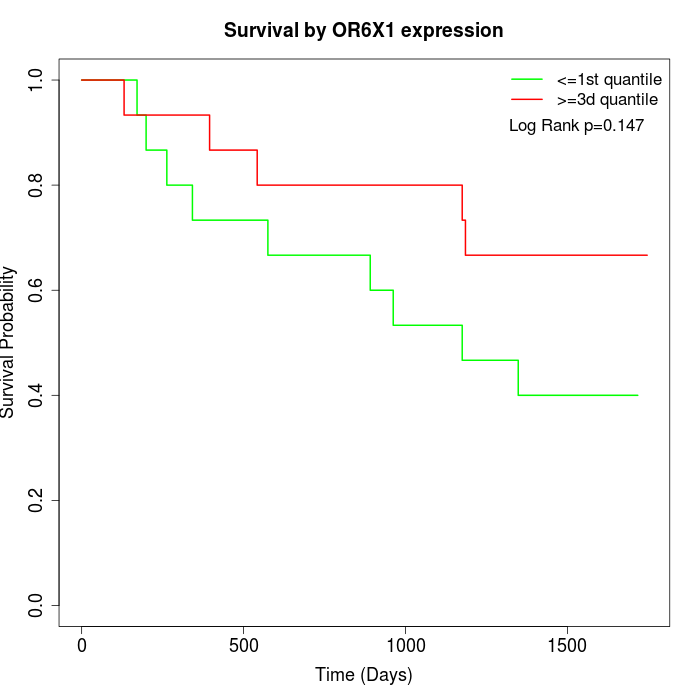
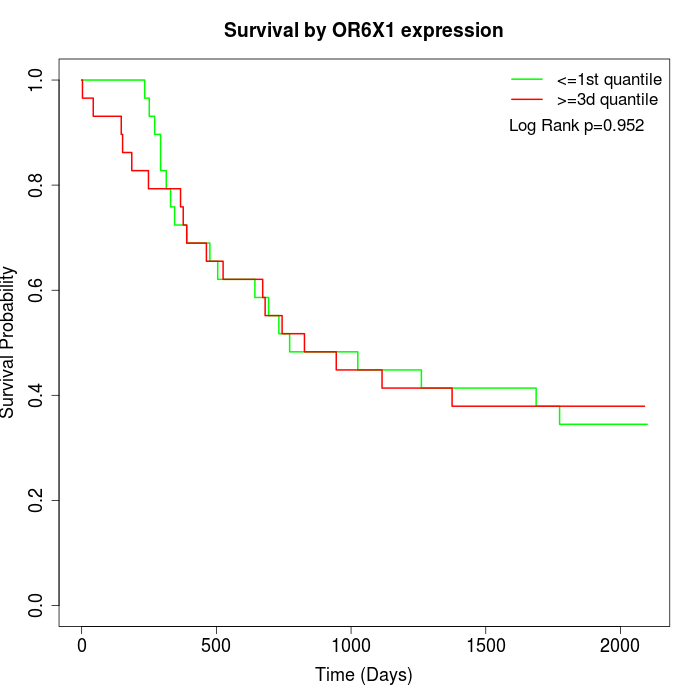
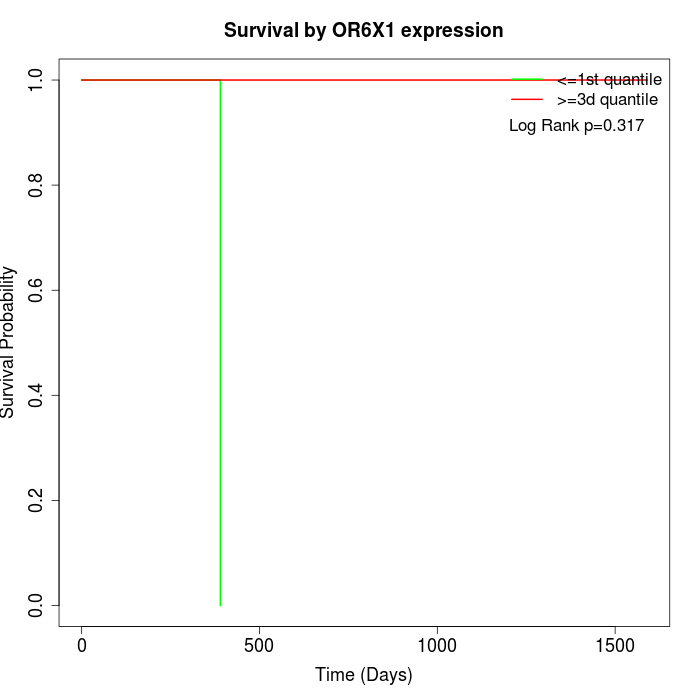
Survival by OR6X1 expression:
Note: Click image to view full size file.
Copy number change of OR6X1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | OR6X1 | 390260 | 0 | 13 | 17 | |
| GSE20123 | OR6X1 | 390260 | 0 | 13 | 17 | |
| GSE43470 | OR6X1 | 390260 | 2 | 7 | 34 | |
| GSE46452 | OR6X1 | 390260 | 3 | 26 | 30 | |
| GSE47630 | OR6X1 | 390260 | 2 | 19 | 19 | |
| GSE54993 | OR6X1 | 390260 | 10 | 0 | 60 | |
| GSE54994 | OR6X1 | 390260 | 5 | 19 | 29 | |
| GSE60625 | OR6X1 | 390260 | 0 | 3 | 8 | |
| GSE74703 | OR6X1 | 390260 | 1 | 5 | 30 | |
| GSE74704 | OR6X1 | 390260 | 0 | 9 | 11 | |
| TCGA | OR6X1 | 390260 | 6 | 50 | 40 |
Total number of gains: 29; Total number of losses: 164; Total Number of normals: 295.
Somatic mutations of OR6X1:
Generating mutation plots.
Highly correlated genes for OR6X1:
Showing all 9 correlated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| OR6X1 | TAS2R39 | 0.751577 | 3 | 0 | 3 |
| OR6X1 | OR2G2 | 0.737119 | 3 | 0 | 3 |
| OR6X1 | OR2Y1 | 0.734619 | 3 | 0 | 3 |
| OR6X1 | TXLNB | 0.702331 | 3 | 0 | 3 |
| OR6X1 | VRTN | 0.682862 | 3 | 0 | 3 |
| OR6X1 | DMC1 | 0.681422 | 3 | 0 | 3 |
| OR6X1 | FAM189A1 | 0.677203 | 3 | 0 | 3 |
| OR6X1 | TDRD9 | 0.674685 | 3 | 0 | 3 |
| OR6X1 | CELF3 | 0.670452 | 3 | 0 | 3 |
For details and further investigation, click here