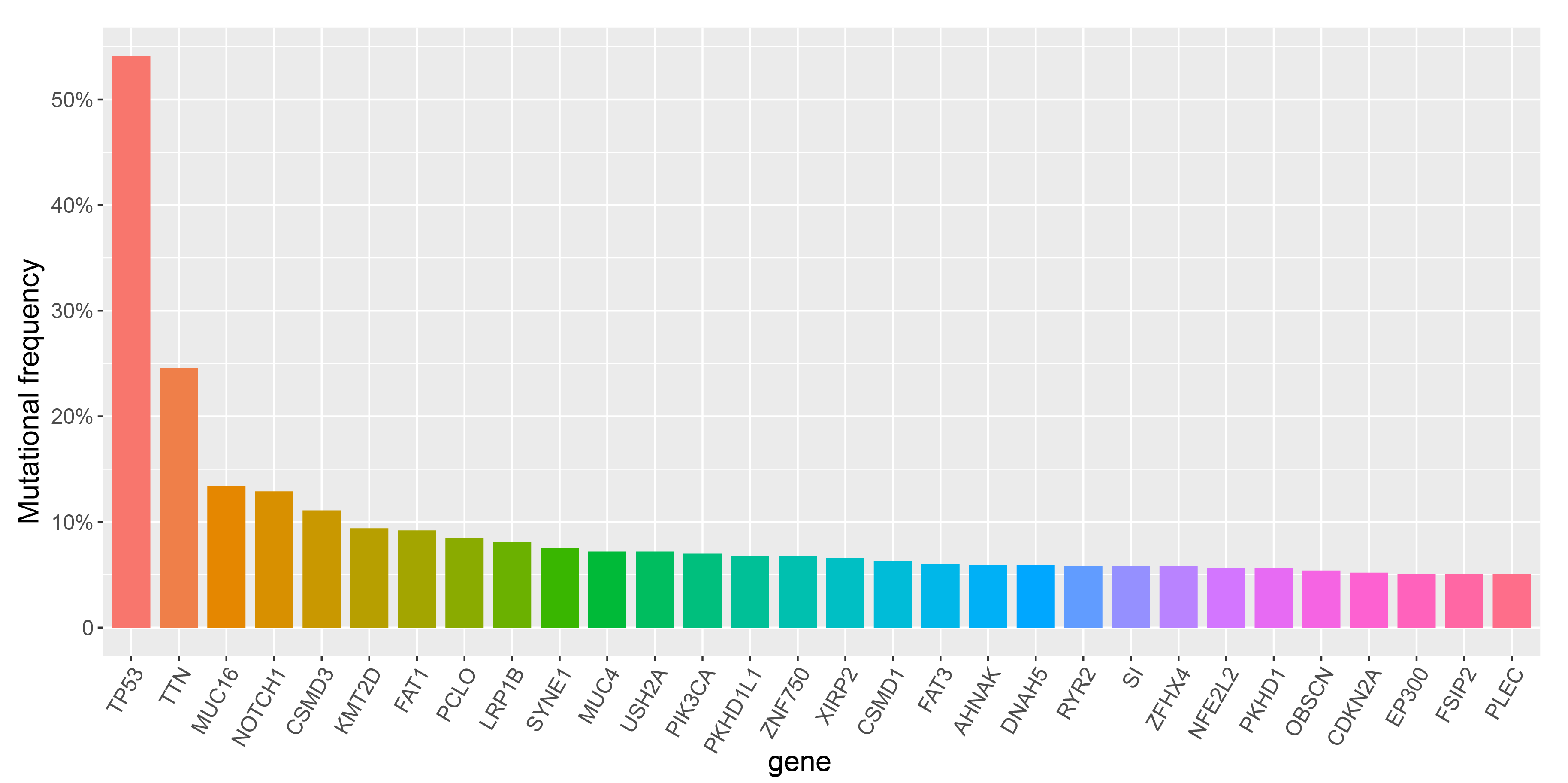
The following genes were mutated in more than 5% of the ESCC samples:
| Gene | No. of Synonymous | No. of Nonsynonymous | Mutational Frequency | Mutation Details | TP53 | 7 | 492 | 54.05% | Show | TTN | 63 | 266 | 24.63% | Show | MUC16 | 43 | 138 | 13.37% | Show | NOTCH1 | 8 | 102 | 12.88% | Show | CSMD3 | 23 | 123 | 11.05% | Show | KMT2D | 10 | 43 | 9.43% | Show | FAT1 | 19 | 35 | 9.22% | Show | PCLO | 26 | 84 | 8.52% | Show | LRP1B | 25 | 86 | 8.09% | Show | SYNE1 | 23 | 80 | 7.53% | Show | MUC4 | 35 | 65 | 7.18% | Show | USH2A | 24 | 68 | 7.18% | Show | PIK3CA | 2 | 97 | 6.97% | Show | PKHD1L1 | 23 | 67 | 6.83% | Show | ZNF750 | 6 | 26 | 6.83% | Show | XIRP2 | 10 | 78 | 6.62% | Show | CSMD1 | 21 | 65 | 6.33% | Show | FAT3 | 23 | 61 | 5.98% | Show | AHNAK | 17 | 63 | 5.91% | Show | DNAH5 | 29 | 50 | 5.91% | Show | RYR2 | 21 | 55 | 5.77% | Show | SI | 11 | 63 | 5.77% | Show | ZFHX4 | 21 | 56 | 5.77% | Show | PKHD1 | 17 | 60 | 5.63% | Show | NFE2L2 | 1 | 75 | 5.56% | Show | OBSCN | 19 | 56 | 5.35% | Show | CDKN2A | 1 | 23 | 5.21% | Show | PLEC | 18 | 50 | 5.14% | Show | EP300 | 3 | 59 | 5.14% | Show | FSIP2 | 6 | 62 | 5.07% | Show |
|---|
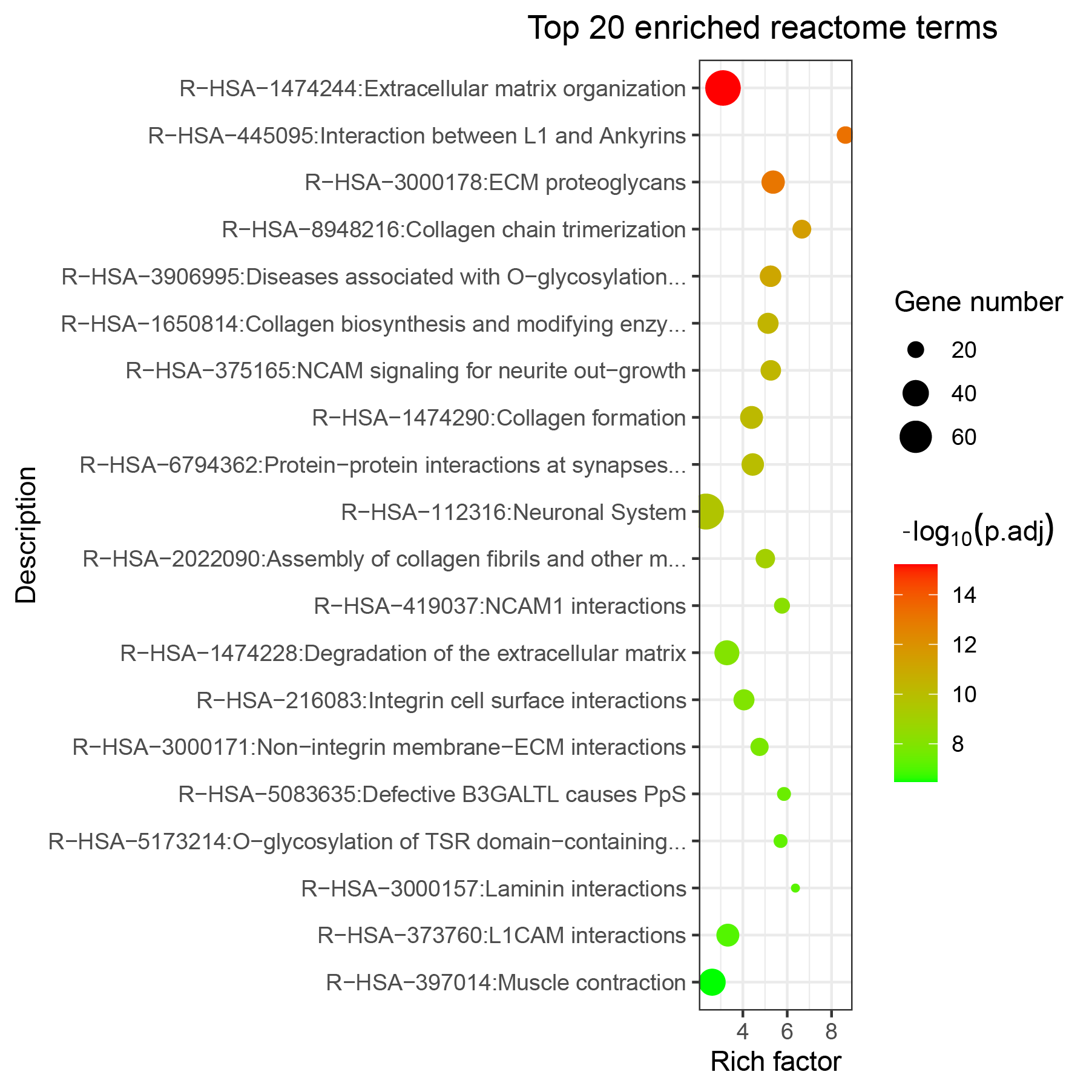
KEGG and Reactome enrichment analysis of genes mutated in more than 1% ESCC samples:

Showing top 1-10/73 KEGG enriched records.
| ID | Description | P.adjust | Count | Genes |
|---|---|---|---|---|
| hsa04512 | ECM-receptor interaction | 0.0000 | 32 | DSPP, RELN, LAMA2, COL6A5, FREM2, COL6A6, LAMA1, VWF, COL6A3, LAMA5, HSPG2, FRAS1, TNR, COL1A2, FREM1, LAMA3, FN1, AGRN, LAMB4, TNXB, COL6A2... |
| hsa04510 | Focal adhesion | 0.0000 | 45 | PIK3CA, RELN, LAMA2, COL6A5, COL6A6, LAMA1, VWF, COL6A3, LAMA5, TNR, COL1A2, TLN2, FLNC, ARHGAP5, LAMA3, FN1, DOCK1, FLT4, EGFR, KDR, MYLK, ... |
| hsa04724 | Glutamatergic synapse | 0.0000 | 32 | CACNA1A, CACNA1C, GNAS, GRIN2B, ADCY2, GRIN3A, ITPR2, KCNJ3, GRIN2A, GRM3, GRIK2, ITPR3, GRIA2, ADCY8, GRM7, GRIK3, ITPR1, ADCY1, GRM1, SHAN... |
| hsa02010 | ABC transporters | 0.0000 | 19 | ABCA13, CFTR, ABCA4, ABCA12, ABCC1, ABCB5, ABCB1, ABCC8, ABCA9, ABCA8, ABCA3, ABCA7, ABCA2, ABCA6, ABCB4, ABCC9, ABCC5, ABCC11, ABCA1 |
| hsa04713 | Circadian entrainment | 0.0000 | 28 | RYR2, RYR1, RYR3, CACNA1I, CACNA1C, GNAS, GRIN2B, ADCY2, KCNJ3, GRIN2A, ITPR3, CACNA1H, GRIA2, ADCY8, ITPR1, ADCY1, CACNA1G, PLCB1, NOS1, GR... |
| hsa05165 | Human papillomavirus infection | 0.0000 | 58 | TP53, NOTCH1, PIK3CA, EP300, RELN, LAMA2, COL6A5, NOTCH3, COL6A6, CREBBP, LAMA1, TBP, VWF, COL6A3, LAMA5, GNAS, TNR, COL1A2, RB1, NOTCH2, AT... |
| hsa04020 | Calcium signaling pathway | 0.0000 | 45 | RYR2, RYR1, RYR3, CACNA1A, CACNA1I, ERBB4, CACNA1C, CACNA1E, GNAS, ADCY2, ITPR2, GRIN2A, CACNA1B, ITPR3, CACNA1H, FLT4, PLCE1, ADCY8, EGFR, ... |
| hsa05017 | Spinocerebellar ataxia | 0.0000 | 32 | PIK3CA, RYR1, RELN, CACNA1A, ATXN1, TBP, GRIN2B, GRIN3A, ITPR2, GRIN2A, ATG2A, ITPR3, SPTBN2, GRIA2, PIK3R4, MTOR, ITPR1, GRM1, ATP2A1, RB1C... |
| hsa04974 | Protein digestion and absorption | 0.0000 | 25 | COL6A5, COL22A1, COL6A6, COL12A1, COL11A1, COL6A3, COL1A2, COL14A1, COL3A1, COL18A1, COL20A1, COL19A1, COL24A1, COL6A2, COL4A1, COL28A1, COL... |
| hsa04720 | Long-term potentiation | 0.0000 | 19 | EP300, CREBBP, CACNA1C, GRIN2B, ITPR2, GRIN2A, ITPR3, GRIA2, ADCY8, RAPGEF3, ITPR1, ADCY1, GRM1, PLCB1, GRIN2C, GRIA1, GRM5, PLCB3, PRKCB |
*double click on genes to view all the genes.
Showing top 1-10/119 Reactome enriched records.
| ID | Description | P.adjust | Count | Genes |
|---|---|---|---|---|
| R-HSA-1474244 | Extracellular matrix organization | 0.0000 | 73 | PLEC, DSPP, LAMA2, COL6A5, DMD, DST, COL22A1, COL6A6, NRXN1, FBN2, COL12A1, VCAN, LAMA1, COL11A1, VWF, COL6A3, LAMA5, HSPG2, TNR, COL1A2, CO... |
| R-HSA-445095 | Interaction between L1 and Ankyrins | 0.0000 | 21 | SPTA1, SCN2A, SCN1A, ANK2, ANK3, SCN9A, SPTB, SPTBN2, SCN4A, SPTBN4, SCN5A, SCN7A, SPTBN5, SCN11A, ANK1, SCN3A, SCN10A, KCNQ3, NFASC, SPTAN1... |
| R-HSA-3000178 | ECM proteoglycans | 0.0000 | 32 | DSPP, LAMA2, COL6A5, COL6A6, VCAN, LAMA1, COL6A3, LAMA5, HSPG2, TNR, COL1A2, COL3A1, ACAN, LAMA3, FN1, AGRN, PTPRS, TNXB, COL6A2, COL4A1, TN... |
| R-HSA-8948216 | Collagen chain trimerization | 0.0000 | 23 | COL6A5, COL22A1, COL6A6, COL12A1, COL11A1, COL6A3, COL1A2, COL14A1, COL3A1, COL18A1, COL20A1, COL19A1, COL24A1, COL6A2, COL4A1, COL28A1, COL... |
| R-HSA-3906995 | Diseases associated with O-glycosylation of proteins | 0.0000 | 28 | MUC16, NOTCH1, MUC4, MUC5B, THSD7A, MUC17, MUC12, NOTCH3, ADAMTS20, NOTCH2, MUC6, ADAMTS16, ADAMTS18, MUC3A, ADAMTS12, ADAMTS5, ADAMTS19, AD... |
| R-HSA-1650814 | Collagen biosynthesis and modifying enzymes | 0.0000 | 27 | COL6A5, COL22A1, COL6A6, COL12A1, COL11A1, COL6A3, COL1A2, COL14A1, COL3A1, COL18A1, COL20A1, COL19A1, COL24A1, COL6A2, COL4A1, COL28A1, TLL... |
| R-HSA-375165 | NCAM signaling for neurite out-growth | 0.0000 | 26 | SPTA1, COL6A5, COL6A6, CACNA1I, CACNA1C, COL6A3, COL3A1, SPTB, SPTBN2, CACNA1H, AGRN, SPTBN4, SPTBN5, CACNA1G, CACNA1S, COL6A2, COL4A1, COL9... |
| R-HSA-1474290 | Collagen formation | 0.0000 | 31 | PLEC, COL6A5, DST, COL22A1, COL6A6, COL12A1, COL11A1, COL6A3, COL1A2, COL14A1, COL3A1, LAMA3, COL18A1, COL20A1, COL19A1, COL24A1, PXDN, COL6... |
| R-HSA-6794362 | Protein-protein interactions at synapses | 0.0000 | 30 | NRXN1, PTPRD, NRXN3, PPFIA2, GRIN2B, GRIN2A, SLITRK3, LRRTM1, SLITRK5, NRXN2, PTPRS, NLGN1, GRM1, SHANK1, SLITRK1, GRIN2C, LRRTM4, GRIA1, LR... |
| R-HSA-112316 | Neuronal System | 0.0000 | 75 | NRXN1, PTPRD, NRXN3, CACNA1A, ERBB4, PPFIA2, CACNA1E, GRIN2B, NBEA, RIMS1, ADCY2, KCND2, GRIN3A, KCNJ3, HCN1, GRIN2A, CACNA1B, SLITRK3, NRG1... |
*double click on genes to view all the genes.