Browse consensus DEGs in ESCCdb
By integrating DEGs from 20 datasets, we identified 789 consensus DEGs that were consistently and significantly differential expressed in more than half of these datasets.
KEGG and reactome enrichment analysis of consensus up-regulated genes
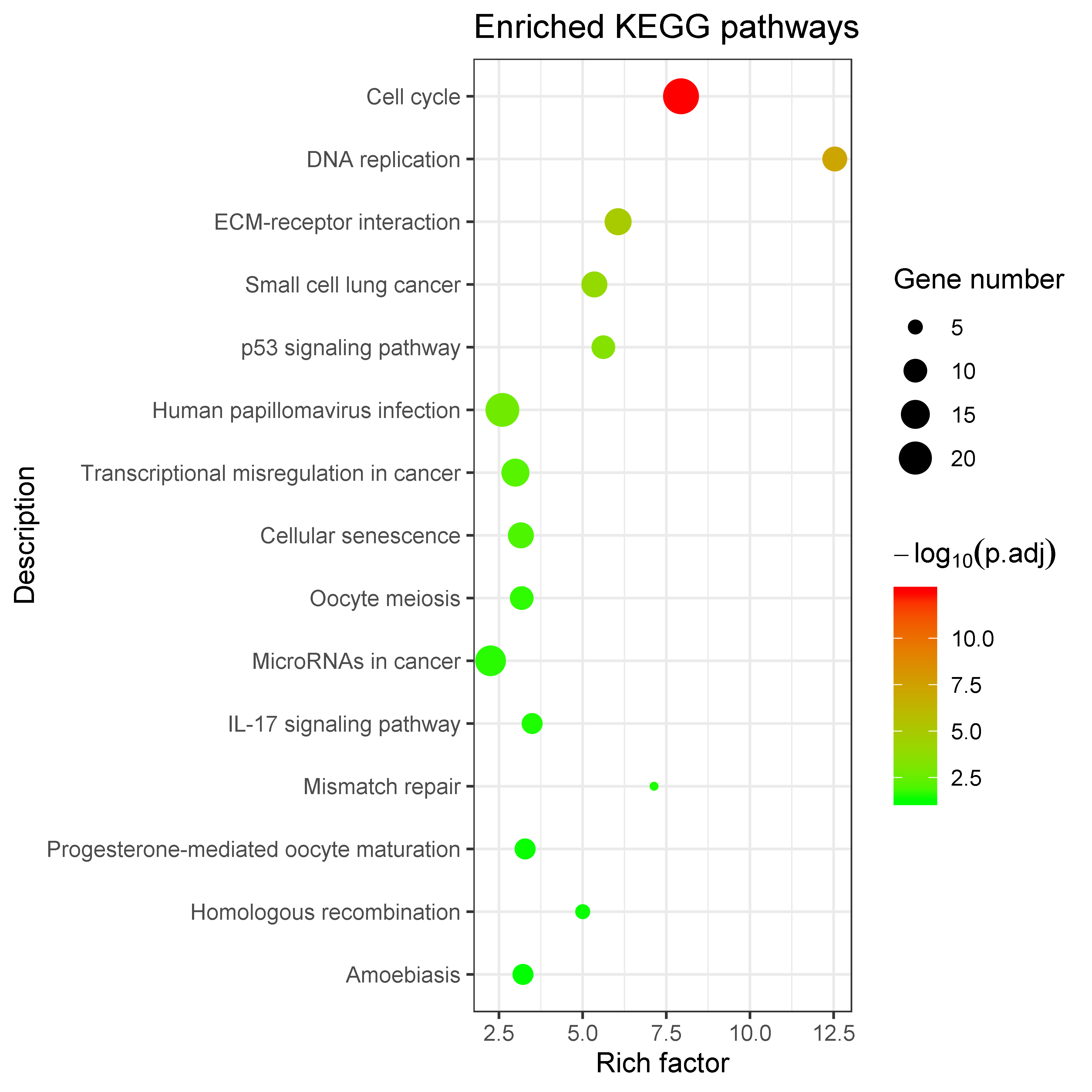

Showing top 1-10/14 KEGG enriched records.
| ID | Description | P.adjust | Count | Genes | Promoter scan |
|---|---|---|---|---|---|
| hsa04110 | Cell cycle | 0.0000 | 24 | MCM6, CDC20, CDC25B, MCM2, ORC6, MAD2L1, BUB1, TTK, CHEK1, MCM5, CDK1, CCNB2, MCM4, MCM7, BUB1B, CDC45, PCNA, CDC6, DBF4, MCM3, CDK4, CCNB1,... | |
| hsa03030 | DNA replication | 0.0000 | 11 | MCM6, MCM2, MCM5, RFC4, MCM4, MCM7, FEN1, PCNA, RNASEH2A, MCM3, PRIM2 | |
| hsa04512 | ECM-receptor interaction | 0.0000 | 13 | LAMC2, LAMB3, ITGB4, AGRN, ITGA6, SPP1, HMMR, COL4A5, COL4A2, ITGAV, LAMA3, TNC, ITGA3 | |
| hsa05222 | Small cell lung cancer | 0.0000 | 12 | CKS1B, LAMC2, LAMB3, ITGA6, CKS2, COL4A5, CDK4, COL4A2, ITGAV, LAMA3, ITGA3, CDK6 | |
| hsa04115 | p53 signaling pathway | 0.0000 | 10 | BID, CHEK1, CDK1, CCNB2, CDK4, CCNB1, IGFBP3, SERPINE1, RRM2, CDK6 | |
| hsa05165 | Human papillomavirus infection | 0.0020 | 21 | LAMC2, LAMB3, ITGB4, FZD6, ITGA6, ATP6V1C1, SPP1, FZD2, HEY1, ISG15, EIF2AK2, COL4A5, CDK4, COL4A2, ITGAV, LAMA3, FADD, TNC, ITGA3, CDK6, WN... | |
| hsa05202 | Transcriptional misregulation in cancer | 0.0080 | 14 | PLAU, SIX4, HOXA10, MLF1, TCF3, CXCL8, MET, MMP9, SIX1, HMGA2, IGFBP3, ETV5, RUNX2, BCL2A1 | |
| hsa04218 | Cellular senescence | 0.0110 | 12 | MYBL2, ITPR3, FOXM1, CHEK1, CDK1, CCNB2, CXCL8, CDK4, CCNB1, IGFBP3, SERPINE1, CDK6 | |
| hsa04114 | Oocyte meiosis | 0.0290 | 10 | ITPR3, CDC20, AURKA, MAD2L1, BUB1, CDK1, CCNB2, FBXO5, CCNB1, PLK1 | |
| hsa05206 | MicroRNAs in cancer | 0.0320 | 17 | PLAU, CDC25B, DNMT3B, CDCA5, KIF23, DNMT1, FSCN1, MET, MMP9, STMN1, HMGA2, TNC, EFNA1, TP63, HOXD10, BRCA1, CDK6 |
Showing top 1-10/100 Reactome enriched records.
| ID | Description | P.adjust | Count | Genes | Promoter scan |
|---|---|---|---|---|---|
| R-HSA-69620 | Cell Cycle Checkpoints | 0.0000 | 42 | UBE2C, MCM6, AURKB, CDC20, MCM2, SPC25, CENPN, EXO1, KIF18A, ORC6, CENPE, CENPF, MAD2L1, BUB1, CENPI, BLM, MCM10, MCM8, CHEK1, CENPA, MCM5, ... | |
| R-HSA-176974 | Unwinding of DNA | 0.0000 | 11 | MCM6, MCM2, MCM8, MCM5, GINS2, GINS1, MCM4, MCM7, CDC45, MCM3, GINS4 | |
| R-HSA-69190 | DNA strand elongation | 0.0000 | 15 | MCM6, MCM2, MCM8, MCM5, RFC4, GINS2, GINS1, MCM4, MCM7, FEN1, CDC45, PCNA, MCM3, GINS4, PRIM2 | |
| R-HSA-453279 | Mitotic G1-G1/S phases | 0.0000 | 27 | MYBL2, CKS1B, GMNN, MCM6, MCM2, ORC6, MCM10, MCM8, MCM5, TOP2A, CDK1, MCM4, MCM7, TK1, CDC45, PCNA, CDC6, FBXO5, DBF4, MCM3, CDK4, CCNB1, RR... | |
| R-HSA-68962 | Activation of the pre-replicative complex | 0.0000 | 15 | GMNN, MCM6, MCM2, ORC6, MCM10, MCM8, MCM5, MCM4, MCM7, CDC45, CDC6, DBF4, MCM3, PRIM2, CDT1 | |
| R-HSA-2500257 | Resolution of Sister Chromatid Cohesion | 0.0000 | 24 | AURKB, CDC20, SPC25, CENPN, KIF18A, CENPE, CDCA5, CENPF, MAD2L1, BUB1, CENPI, CENPA, NUF2, CDCA8, KIF2C, CDK1, CCNB2, NUP107, NDC80, BUB1B, ... | |
| R-HSA-69206 | G1/S Transition | 0.0000 | 24 | CKS1B, GMNN, MCM6, MCM2, ORC6, MCM10, MCM8, MCM5, CDK1, MCM4, MCM7, TK1, CDC45, PCNA, CDC6, FBXO5, DBF4, MCM3, CDK4, CCNB1, RRM2, TYMS, PRIM... | |
| R-HSA-68877 | Mitotic Prometaphase | 0.0000 | 28 | AURKB, CDC20, SPC25, NEK2, CENPN, KIF18A, CENPE, CDCA5, CENPF, MAD2L1, BUB1, CENPI, CENPA, NUF2, NCAPH, CDCA8, NCAPG, KIF2C, CDK1, CCNB2, NU... | |
| R-HSA-176187 | Activation of ATR in response to replication stress | 0.0000 | 14 | MCM6, MCM2, ORC6, MCM10, MCM8, CHEK1, MCM5, RFC4, MCM4, MCM7, CDC45, CDC6, DBF4, MCM3 | |
| R-HSA-141424 | Amplification of signal from the kinetochores | 0.0000 | 20 | AURKB, CDC20, SPC25, CENPN, KIF18A, CENPE, CENPF, MAD2L1, BUB1, CENPI, CENPA, NUF2, CDCA8, KIF2C, NUP107, NDC80, BUB1B, BIRC5, KNTC1, PLK1 |
Full KEGG and Reactome enrichment results is available here
KEGG and reactome enrichment analysis of consensus down-regulated genes

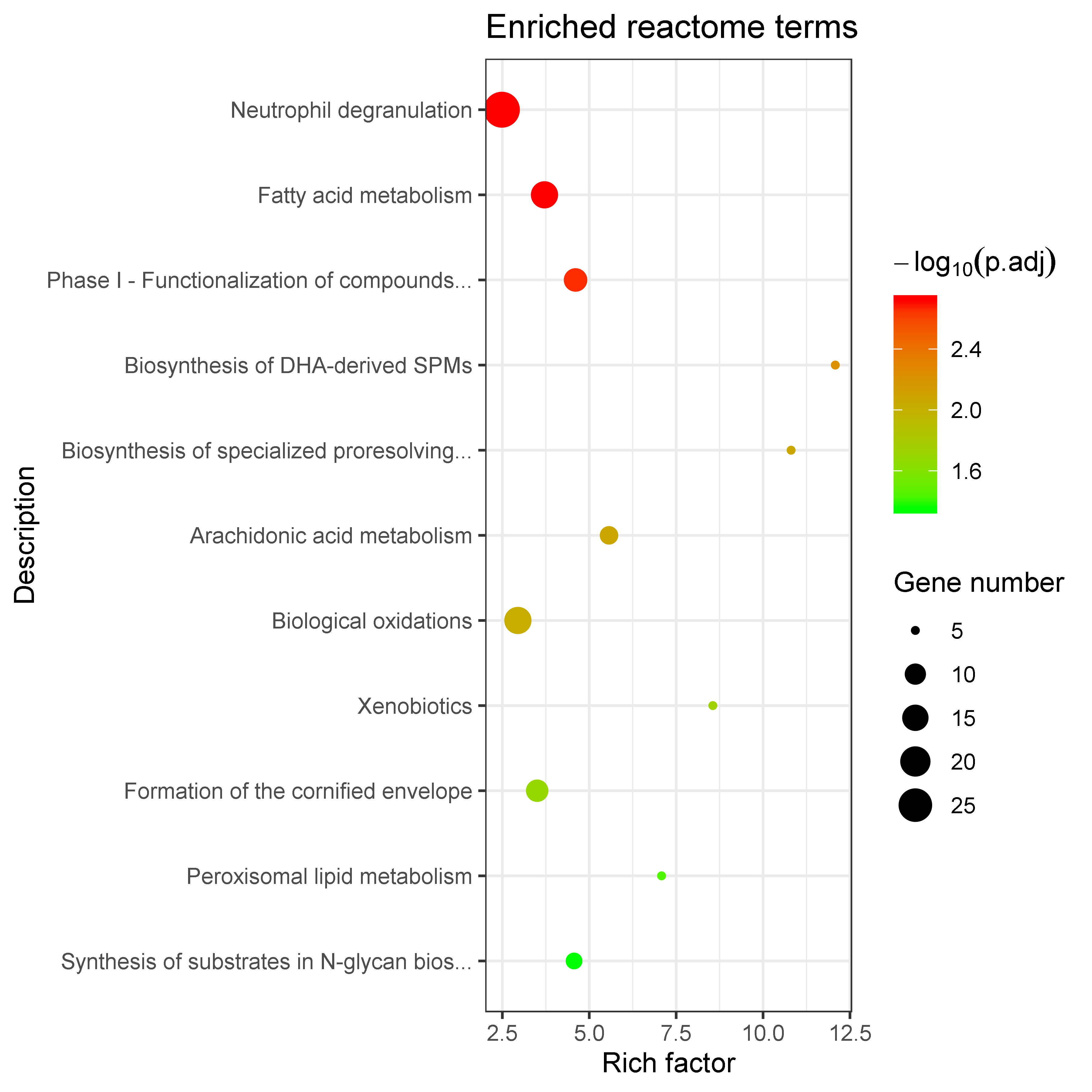
Showing all 10 KEGG enriched records.
| ID | Description | P.adjust | Count | Genes | Promoter scan |
|---|---|---|---|---|---|
| hsa00071 | Fatty acid degradation | 0.0000 | 9 | ALDH3A2, ADH1B, ACOX3, ACOX1, ACAA1, ALDH9A1, ACADM, ADH7, ALDH7A1 | |
| hsa00590 | Arachidonic acid metabolism | 0.0010 | 9 | CBR3, GPX3, EPHX2, CYP2J2, ALOX12, CYP2C9, CYP2E1, PTGS1, ALOX15B | |
| hsa00410 | beta-Alanine metabolism | 0.0040 | 6 | ALDH3A2, ACOX3, ACOX1, ALDH9A1, ABAT, ALDH7A1 | |
| hsa01040 | Biosynthesis of unsaturated fatty acids | 0.0200 | 5 | ACOX3, ACOX1, ACAA1, ELOVL6, ELOVL4 | |
| hsa00280 | Valine, leucine and isoleucine degradation | 0.0350 | 6 | ALDH3A2, ACAA1, ALDH9A1, ACADM, ABAT, ALDH7A1 | |
| hsa00830 | Retinol metabolism | 0.0350 | 7 | ADH1B, CYP2C18, CYP2C9, DHRS9, ADH7, CYP3A5, RDH12 | |
| hsa04726 | Serotonergic synapse | 0.0350 | 9 | CYP2J2, ITPR2, CYP2C18, ALOX12, CYP2C9, PTGS1, DUSP1, ALOX15B, MAPK3 | |
| hsa00982 | Drug metabolism - cytochrome P450 | 0.0350 | 7 | MGST2, ADH1B, FMO2, CYP2C9, CYP2E1, ADH7, CYP3A5 | |
| hsa01212 | Fatty acid metabolism | 0.0410 | 6 | ACOX3, ACOX1, ACAA1, ELOVL6, ELOVL4, ACADM | |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 0.0410 | 7 | CBR3, MGST2, ADH1B, CYP2C9, CYP2E1, ADH7, CYP3A5 |
Showing top 1-10/11 Reactome enriched records.
| ID | Description | P.adjust | Count | Genes | Promoter scan |
|---|---|---|---|---|---|
| R-HSA-6798695 | Neutrophil degranulation | 0.0020 | 29 | CRISP3, CFD, GSN, ACPP, SYNGR1, METTL7A, CXCR2, VAT1, HEBP2, TOM1, PRSS3, ACAA1, SERPINB1, CPPED1, CEACAM1, NBEAL2, CSTB, CDA, SERPINB6, ATP... | |
| R-HSA-8978868 | Fatty acid metabolism | 0.0020 | 16 | CYP4B1, ACOX2, EPHX2, ALDH3A2, ACOX3, CYP2J2, ACOX1, ACAA1, HPGD, ELOVL6, ALOX12, CYP2C9, ELOVL4, ACADM, PTGS1, ALOX15B | |
| R-HSA-211945 | Phase I - Functionalization of compounds | 0.0020 | 12 | CBR3, CYP4B1, ADH1B, FMO2, CYP2J2, CYP2C18, CES2, CYP2C9, CYP2E1, PTGS1, ADH7, CYP3A5 | |
| R-HSA-9018677 | Biosynthesis of DHA-derived SPMs | 0.0060 | 5 | EPHX2, HPGD, ALOX12, CYP2C9, CYP2E1 | |
| R-HSA-9018678 | Biosynthesis of specialized proresolving mediators (SPMs) | 0.0080 | 5 | EPHX2, HPGD, ALOX12, CYP2C9, CYP2E1 | |
| R-HSA-2142753 | Arachidonic acid metabolism | 0.0080 | 8 | CYP4B1, EPHX2, CYP2J2, HPGD, ALOX12, CYP2C9, PTGS1, ALOX15B | |
| R-HSA-211859 | Biological oxidations | 0.0090 | 16 | CBR3, CYP4B1, MGST2, ADH1B, FMO2, CYP2J2, SULT2B1, CYP2C18, CES2, CYP2C9, CYP2E1, PTGS1, ADH7, CYP3A5, SLC26A2, CHAC1 | |
| R-HSA-211981 | Xenobiotics | 0.0180 | 5 | CYP2J2, CYP2C18, CYP2C9, CYP2E1, CYP3A5 | |
| R-HSA-6809371 | Formation of the cornified envelope | 0.0200 | 11 | SPINK5, KRT78, EVPL, TGM1, KLK13, FLG, KLK12, IVL, KRT13, SPRR3, SPRR1A | |
| R-HSA-390918 | Peroxisomal lipid metabolism | 0.0360 | 5 | ACOX2, ALDH3A2, ACOX3, ACOX1, ACAA1 |