| Full name: NEDD4 binding protein 2 | Alias Symbol: B3BP | ||
| Type: protein-coding gene | Cytoband: 4p14 | ||
| Entrez ID: 55728 | HGNC ID: HGNC:29851 | Ensembl Gene: ENSG00000078177 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of N4BP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | N4BP2 | 55728 | 228242_at | 0.3715 | 0.6393 | |
| GSE26886 | N4BP2 | 55728 | 228242_at | 0.1455 | 0.6361 | |
| GSE45670 | N4BP2 | 55728 | 228242_at | 0.0416 | 0.8745 | |
| GSE53622 | N4BP2 | 55728 | 23706 | -0.0085 | 0.9185 | |
| GSE53624 | N4BP2 | 55728 | 23706 | -0.0867 | 0.2450 | |
| GSE63941 | N4BP2 | 55728 | 228242_at | -0.6187 | 0.4711 | |
| GSE77861 | N4BP2 | 55728 | 228242_at | 0.0351 | 0.9199 | |
| GSE97050 | N4BP2 | 55728 | A_23_P309779 | 0.0224 | 0.9206 | |
| SRP007169 | N4BP2 | 55728 | RNAseq | -0.0420 | 0.9031 | |
| SRP008496 | N4BP2 | 55728 | RNAseq | 0.1337 | 0.5850 | |
| SRP064894 | N4BP2 | 55728 | RNAseq | -0.3584 | 0.2128 | |
| SRP133303 | N4BP2 | 55728 | RNAseq | 0.0218 | 0.9035 | |
| SRP159526 | N4BP2 | 55728 | RNAseq | 0.1142 | 0.6115 | |
| SRP193095 | N4BP2 | 55728 | RNAseq | -0.2521 | 0.0096 | |
| SRP219564 | N4BP2 | 55728 | RNAseq | -0.1018 | 0.7650 | |
| TCGA | N4BP2 | 55728 | RNAseq | 0.0056 | 0.9463 |
Upregulated datasets: 0; Downregulated datasets: 0.
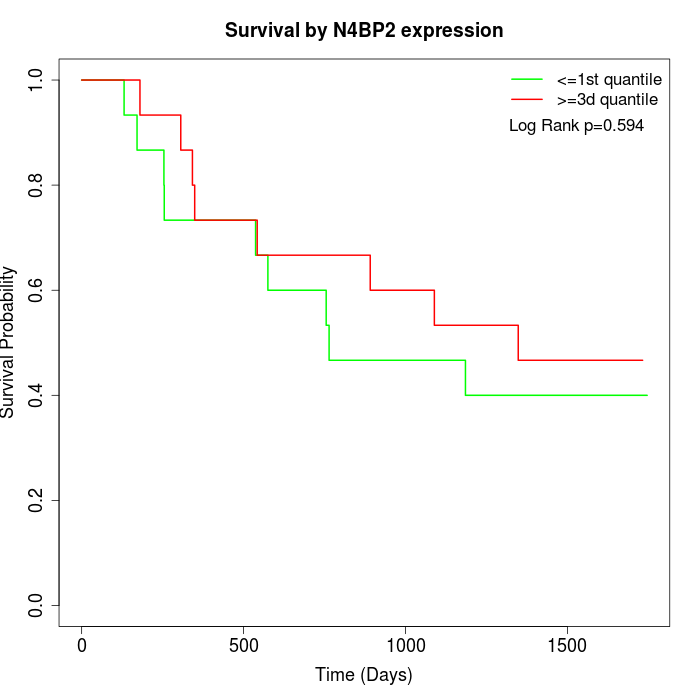
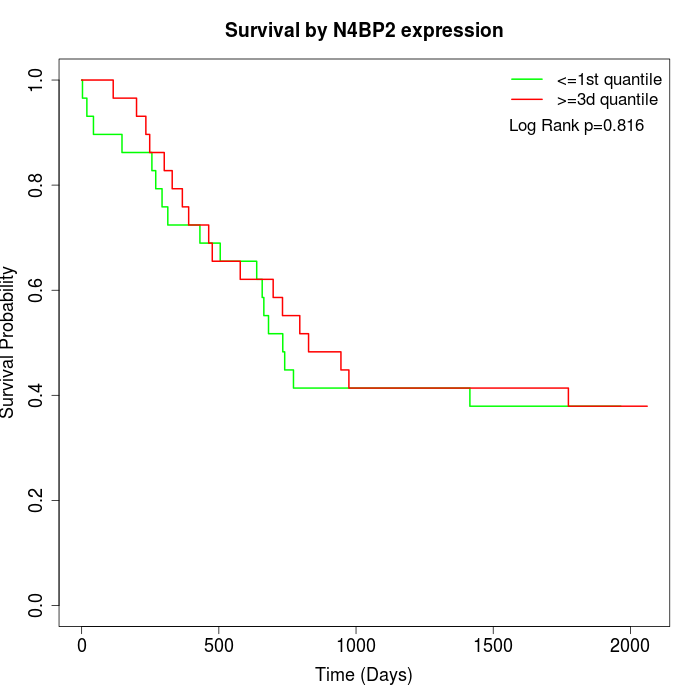
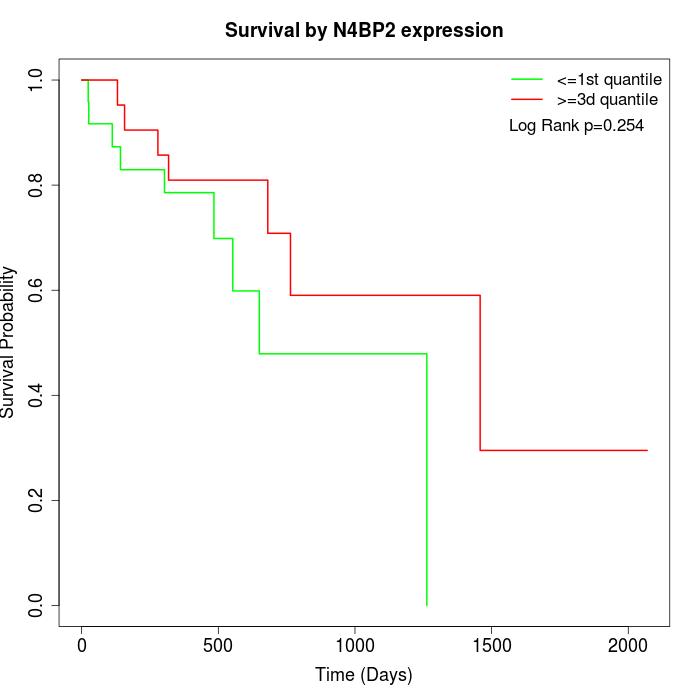
Survival by N4BP2 expression:
Note: Click image to view full size file.
Copy number change of N4BP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | N4BP2 | 55728 | 0 | 16 | 14 | |
| GSE20123 | N4BP2 | 55728 | 0 | 16 | 14 | |
| GSE43470 | N4BP2 | 55728 | 0 | 15 | 28 | |
| GSE46452 | N4BP2 | 55728 | 1 | 36 | 22 | |
| GSE47630 | N4BP2 | 55728 | 1 | 19 | 20 | |
| GSE54993 | N4BP2 | 55728 | 7 | 0 | 63 | |
| GSE54994 | N4BP2 | 55728 | 3 | 10 | 40 | |
| GSE60625 | N4BP2 | 55728 | 0 | 0 | 11 | |
| GSE74703 | N4BP2 | 55728 | 0 | 12 | 24 | |
| GSE74704 | N4BP2 | 55728 | 0 | 9 | 11 | |
| TCGA | N4BP2 | 55728 | 10 | 45 | 41 |
Total number of gains: 22; Total number of losses: 178; Total Number of normals: 288.
Somatic mutations of N4BP2:
Generating mutation plots.
Highly correlated genes for N4BP2:
Showing all 8 correlated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| N4BP2 | DTWD2 | 0.633374 | 4 | 0 | 3 |
| N4BP2 | KLHDC4 | 0.631686 | 3 | 0 | 3 |
| N4BP2 | KATNAL2 | 0.629429 | 3 | 0 | 3 |
| N4BP2 | SLC2A6 | 0.611028 | 3 | 0 | 3 |
| N4BP2 | MBTD1 | 0.598735 | 5 | 0 | 4 |
| N4BP2 | JRKL | 0.578179 | 3 | 0 | 3 |
| N4BP2 | TANK | 0.552393 | 5 | 0 | 3 |
| N4BP2 | LCORL | 0.501573 | 5 | 0 | 3 |
For details and further investigation, click here