| Full name: testis expressed 36 | Alias Symbol: bA383C5.1 | ||
| Type: protein-coding gene | Cytoband: 10q26.13 | ||
| Entrez ID: 387718 | HGNC ID: HGNC:31653 | Ensembl Gene: ENSG00000175018 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of TEX36:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TEX36 | 387718 | 244789_at | 0.0176 | 0.9518 | |
| GSE26886 | TEX36 | 387718 | 244789_at | -0.0417 | 0.7073 | |
| GSE45670 | TEX36 | 387718 | 244789_at | 0.0322 | 0.6707 | |
| GSE53622 | TEX36 | 387718 | 25123 | 0.0876 | 0.5724 | |
| GSE53624 | TEX36 | 387718 | 25123 | 0.1798 | 0.0996 | |
| GSE63941 | TEX36 | 387718 | 244789_at | 0.2814 | 0.6054 | |
| GSE77861 | TEX36 | 387718 | 244789_at | -0.0561 | 0.5303 |
Upregulated datasets: 0; Downregulated datasets: 0.
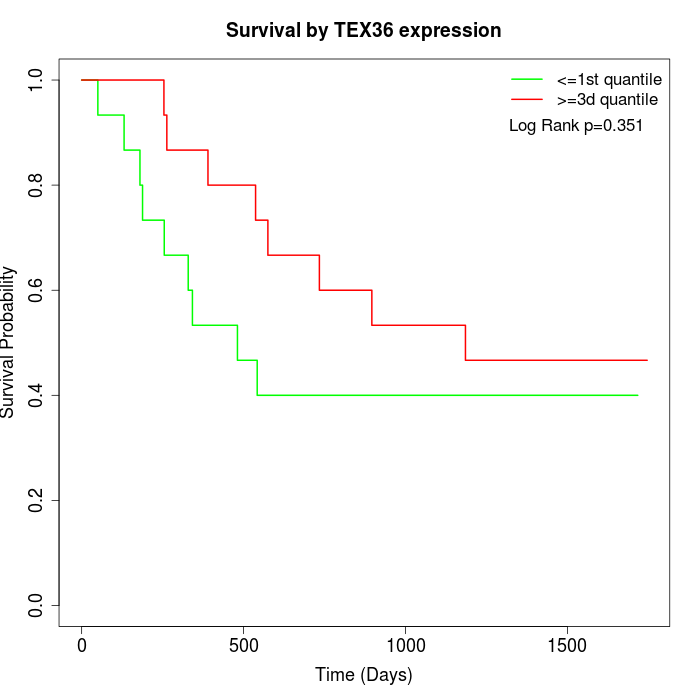
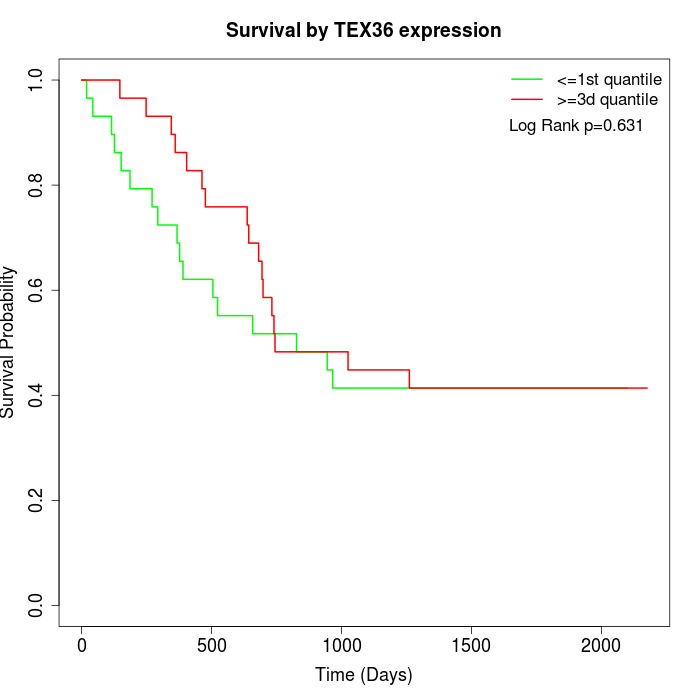
Survival by TEX36 expression:
Note: Click image to view full size file.
Copy number change of TEX36:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TEX36 | 387718 | 2 | 6 | 22 | |
| GSE20123 | TEX36 | 387718 | 2 | 6 | 22 | |
| GSE43470 | TEX36 | 387718 | 0 | 10 | 33 | |
| GSE46452 | TEX36 | 387718 | 0 | 11 | 48 | |
| GSE47630 | TEX36 | 387718 | 2 | 14 | 24 | |
| GSE54993 | TEX36 | 387718 | 8 | 1 | 61 | |
| GSE54994 | TEX36 | 387718 | 1 | 9 | 43 | |
| GSE60625 | TEX36 | 387718 | 0 | 0 | 11 | |
| GSE74703 | TEX36 | 387718 | 0 | 6 | 30 | |
| GSE74704 | TEX36 | 387718 | 2 | 2 | 16 | |
| TCGA | TEX36 | 387718 | 7 | 27 | 62 |
Total number of gains: 24; Total number of losses: 92; Total Number of normals: 372.
Somatic mutations of TEX36:
Generating mutation plots.
Highly correlated genes for TEX36:
Showing all 6 correlated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TEX36 | CCDC30 | 0.565102 | 4 | 0 | 3 |
| TEX36 | CUBN | 0.557139 | 3 | 0 | 3 |
| TEX36 | GMNC | 0.557028 | 3 | 0 | 3 |
| TEX36 | SMIM2 | 0.553255 | 3 | 0 | 3 |
| TEX36 | MYOD1 | 0.539949 | 4 | 0 | 3 |
| TEX36 | DMWD | 0.530174 | 4 | 0 | 3 |
For details and further investigation, click here