| Full name: WNK lysine deficient protein kinase 3 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xp11.22 | ||
| Entrez ID: 65267 | HGNC ID: HGNC:14543 | Ensembl Gene: ENSG00000196632 | OMIM ID: 300358 |
| Drug and gene relationship at DGIdb | |||
Expression of WNK3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | WNK3 | 65267 | 236236_at | 1.0048 | 0.2692 | |
| GSE26886 | WNK3 | 65267 | 1555248_a_at | -0.1384 | 0.2787 | |
| GSE45670 | WNK3 | 65267 | 236236_at | -0.1021 | 0.6768 | |
| GSE53622 | WNK3 | 65267 | 20034 | -0.2125 | 0.2700 | |
| GSE53624 | WNK3 | 65267 | 13030 | -0.0824 | 0.5357 | |
| GSE63941 | WNK3 | 65267 | 236236_at | -0.9711 | 0.0164 | |
| GSE77861 | WNK3 | 65267 | 236236_at | -0.1569 | 0.1869 | |
| SRP007169 | WNK3 | 65267 | RNAseq | 0.7673 | 0.3864 | |
| SRP064894 | WNK3 | 65267 | RNAseq | -0.3015 | 0.2800 | |
| SRP133303 | WNK3 | 65267 | RNAseq | -0.0004 | 0.9991 | |
| SRP159526 | WNK3 | 65267 | RNAseq | 1.1641 | 0.1655 | |
| SRP193095 | WNK3 | 65267 | RNAseq | -0.1046 | 0.6516 | |
| SRP219564 | WNK3 | 65267 | RNAseq | -0.1557 | 0.7296 | |
| TCGA | WNK3 | 65267 | RNAseq | -0.7197 | 0.0614 |
Upregulated datasets: 0; Downregulated datasets: 0.
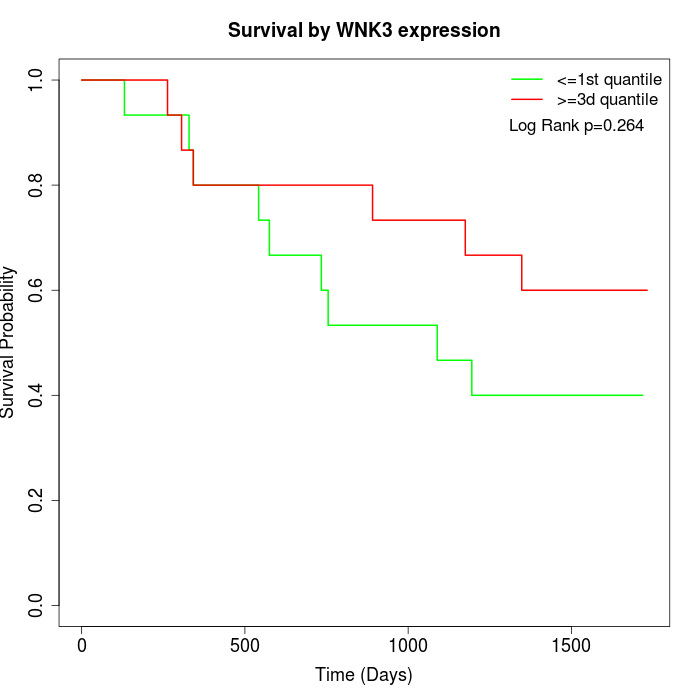
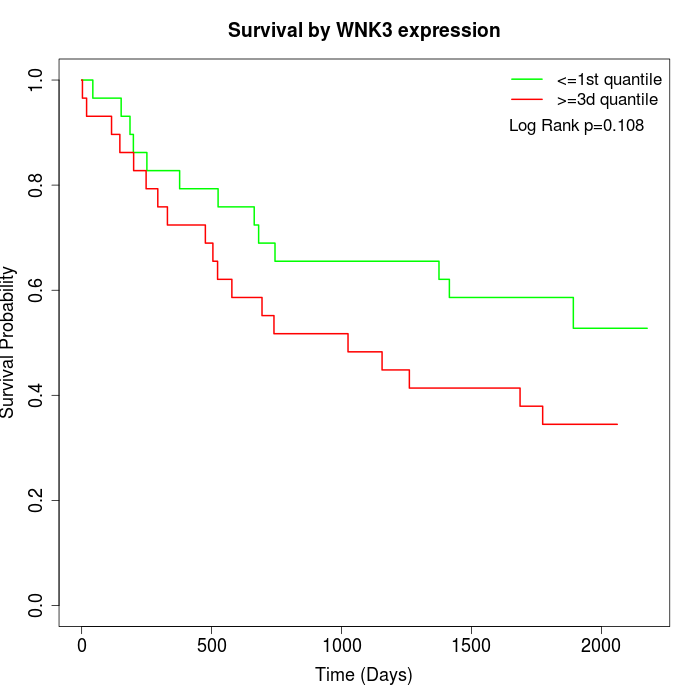
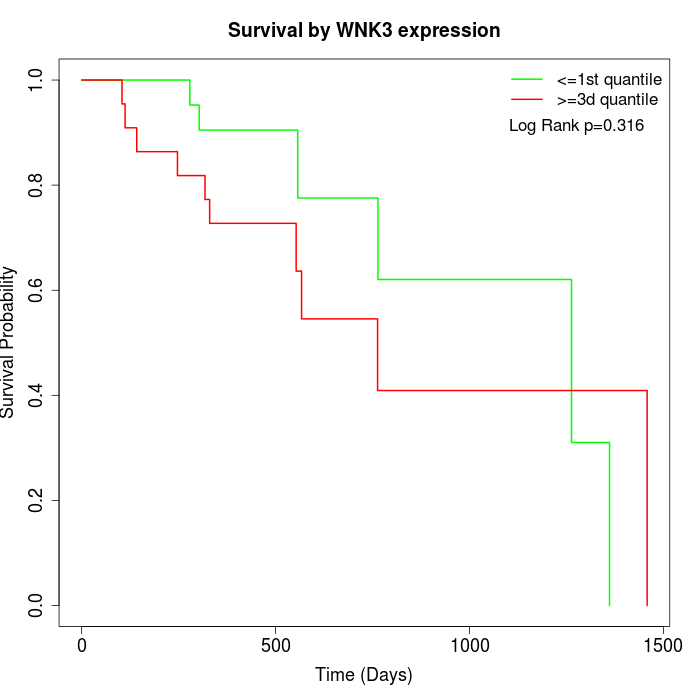
Survival by WNK3 expression:
Note: Click image to view full size file.
Copy number change of WNK3:
No record found for this gene.
Somatic mutations of WNK3:
Generating mutation plots.
Highly correlated genes for WNK3:
Showing all 12 correlated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| WNK3 | PABPC4L | 0.664504 | 3 | 0 | 3 |
| WNK3 | NECAP2 | 0.587169 | 3 | 0 | 3 |
| WNK3 | ZCCHC18 | 0.582066 | 3 | 0 | 3 |
| WNK3 | PDE10A | 0.574476 | 3 | 0 | 3 |
| WNK3 | UGT2B28 | 0.573279 | 3 | 0 | 3 |
| WNK3 | CACNA2D2 | 0.57203 | 5 | 0 | 3 |
| WNK3 | CAMK4 | 0.560287 | 4 | 0 | 3 |
| WNK3 | OR11A1 | 0.532502 | 4 | 0 | 3 |
| WNK3 | COL9A2 | 0.528089 | 4 | 0 | 3 |
| WNK3 | ZNF439 | 0.528082 | 5 | 0 | 3 |
| WNK3 | STS | 0.526837 | 4 | 0 | 3 |
| WNK3 | KCTD16 | 0.52275 | 3 | 0 | 3 |
For details and further investigation, click here