| Full name: potassium channel tetramerization domain containing 16 | Alias Symbol: KIAA1317 | ||
| Type: protein-coding gene | Cytoband: 5q31.3 | ||
| Entrez ID: 57528 | HGNC ID: HGNC:29244 | Ensembl Gene: ENSG00000183775 | OMIM ID: 613423 |
| Drug and gene relationship at DGIdb | |||
Expression of KCTD16:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCTD16 | 57528 | 233234_at | -0.1330 | 0.5347 | |
| GSE26886 | KCTD16 | 57528 | 233234_at | 0.0506 | 0.6639 | |
| GSE45670 | KCTD16 | 57528 | 233234_at | 0.0369 | 0.6725 | |
| GSE53622 | KCTD16 | 57528 | 50024 | 0.1213 | 0.2740 | |
| GSE53624 | KCTD16 | 57528 | 50024 | -0.2205 | 0.0981 | |
| GSE63941 | KCTD16 | 57528 | 233234_at | -1.9544 | 0.0000 | |
| GSE77861 | KCTD16 | 57528 | 233234_at | -0.1608 | 0.1055 | |
| SRP133303 | KCTD16 | 57528 | RNAseq | 0.2197 | 0.5074 | |
| TCGA | KCTD16 | 57528 | RNAseq | -0.2761 | 0.5665 |
Upregulated datasets: 0; Downregulated datasets: 1.
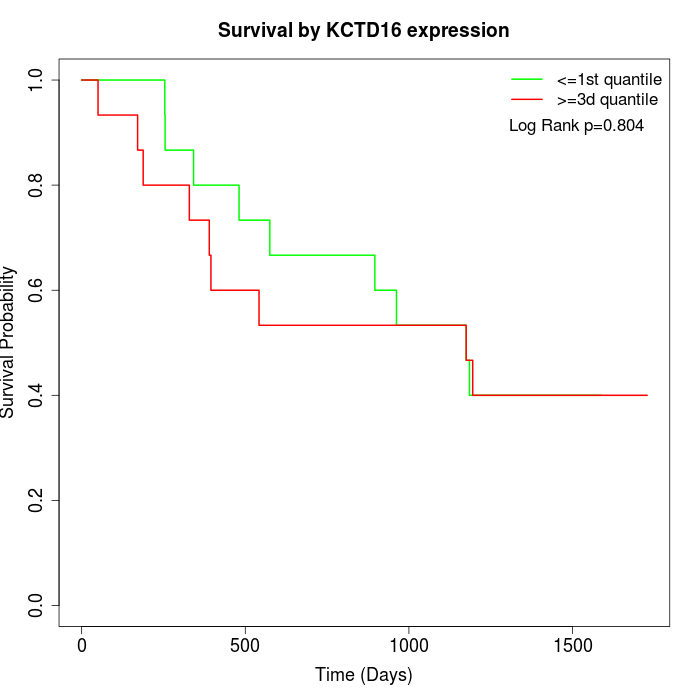
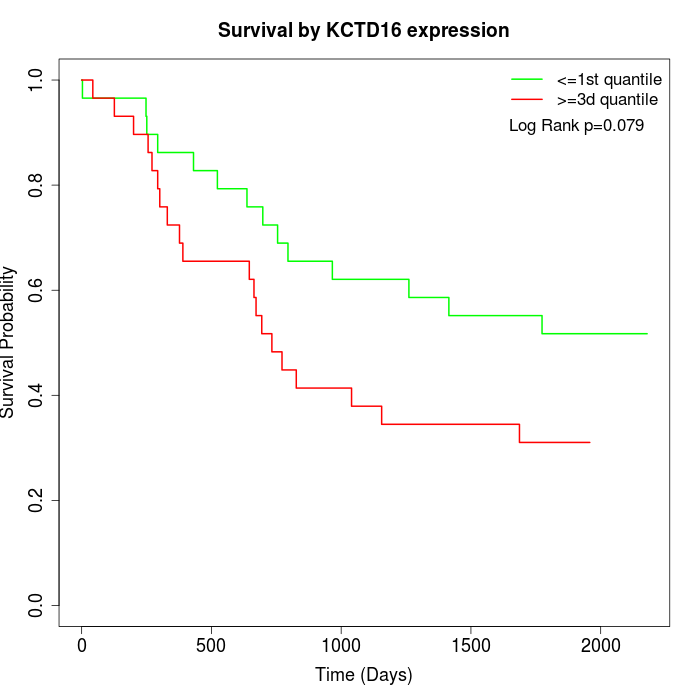
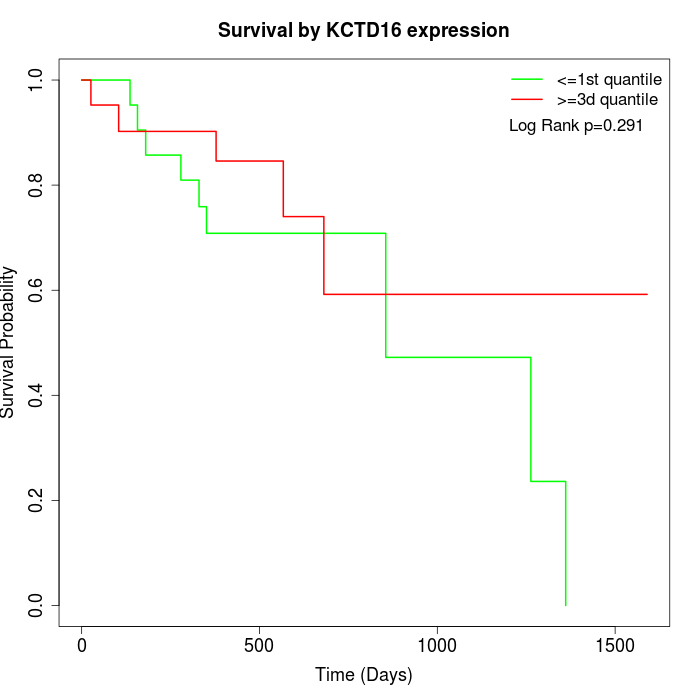
Survival by KCTD16 expression:
Note: Click image to view full size file.
Copy number change of KCTD16:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCTD16 | 57528 | 1 | 11 | 18 | |
| GSE20123 | KCTD16 | 57528 | 1 | 11 | 18 | |
| GSE43470 | KCTD16 | 57528 | 2 | 9 | 32 | |
| GSE46452 | KCTD16 | 57528 | 0 | 27 | 32 | |
| GSE47630 | KCTD16 | 57528 | 0 | 20 | 20 | |
| GSE54993 | KCTD16 | 57528 | 9 | 1 | 60 | |
| GSE54994 | KCTD16 | 57528 | 2 | 14 | 37 | |
| GSE60625 | KCTD16 | 57528 | 0 | 0 | 11 | |
| GSE74703 | KCTD16 | 57528 | 2 | 7 | 27 | |
| GSE74704 | KCTD16 | 57528 | 1 | 6 | 13 | |
| TCGA | KCTD16 | 57528 | 3 | 38 | 55 |
Total number of gains: 21; Total number of losses: 144; Total Number of normals: 323.
Somatic mutations of KCTD16:
Generating mutation plots.
Highly correlated genes for KCTD16:
Showing all 15 correlated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCTD16 | ACOX2 | 0.637437 | 3 | 0 | 3 |
| KCTD16 | LINC01398 | 0.619144 | 3 | 0 | 3 |
| KCTD16 | BRINP1 | 0.619005 | 3 | 0 | 3 |
| KCTD16 | MKRN2 | 0.61359 | 3 | 0 | 3 |
| KCTD16 | MOSPD2 | 0.61295 | 3 | 0 | 3 |
| KCTD16 | NPAS2 | 0.606073 | 3 | 0 | 3 |
| KCTD16 | KCTD6 | 0.59297 | 3 | 0 | 3 |
| KCTD16 | MAPK7 | 0.5698 | 4 | 0 | 3 |
| KCTD16 | BCAP29 | 0.539755 | 3 | 0 | 3 |
| KCTD16 | TMEM234 | 0.523819 | 4 | 0 | 3 |
| KCTD16 | NEGR1-IT1 | 0.522859 | 4 | 0 | 3 |
| KCTD16 | WNK3 | 0.52275 | 3 | 0 | 3 |
| KCTD16 | SLC30A9 | 0.520031 | 4 | 0 | 3 |
| KCTD16 | LINC00922 | 0.516342 | 5 | 0 | 3 |
| KCTD16 | ANO4 | 0.514673 | 4 | 0 | 3 |
For details and further investigation, click here