| Full name: AT-rich interaction domain 3C | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 9p13.3 | ||
| Entrez ID: 138715 | HGNC ID: HGNC:21209 | Ensembl Gene: ENSG00000205143 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of ARID3C:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | ARID3C | 138715 | 19086 | 0.1447 | 0.3265 | |
| GSE53624 | ARID3C | 138715 | 19086 | -0.4605 | 0.0008 | |
| GSE97050 | ARID3C | 138715 | A_33_P3424187 | 0.3818 | 0.3660 | |
| TCGA | ARID3C | 138715 | RNAseq | 1.1867 | 0.1778 |
Upregulated datasets: 0; Downregulated datasets: 0.
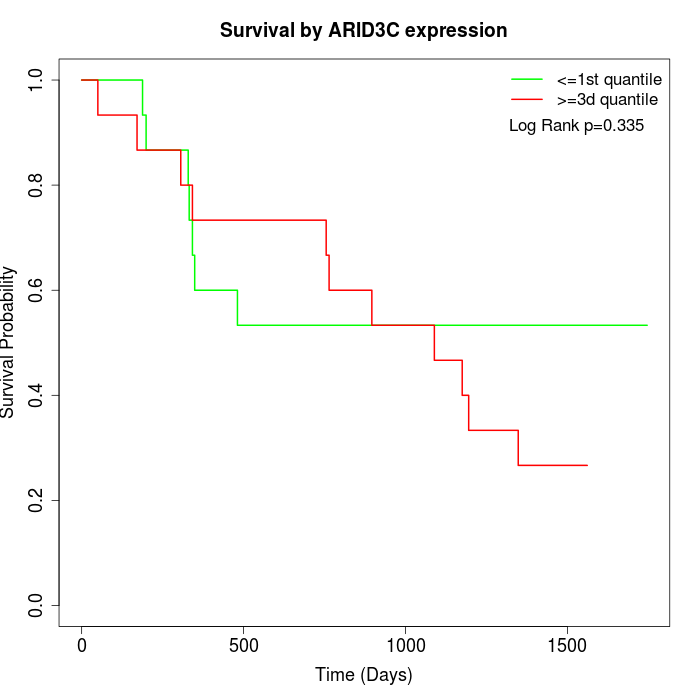
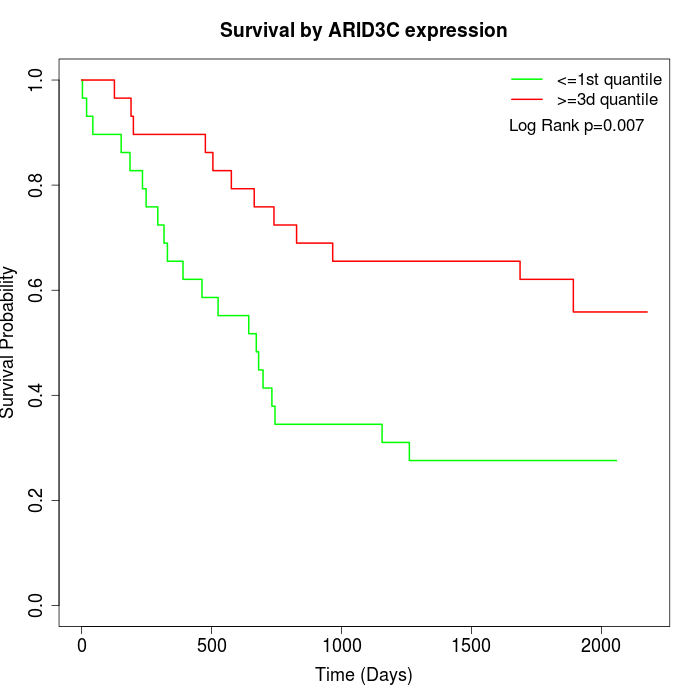
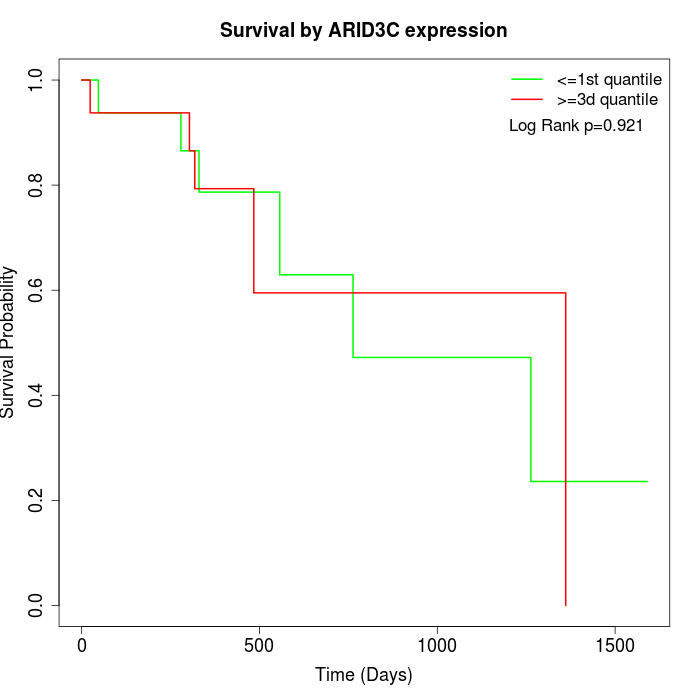
Survival by ARID3C expression:
Note: Click image to view full size file.
Copy number change of ARID3C:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ARID3C | 138715 | 4 | 13 | 13 | |
| GSE20123 | ARID3C | 138715 | 4 | 13 | 13 | |
| GSE43470 | ARID3C | 138715 | 3 | 10 | 30 | |
| GSE46452 | ARID3C | 138715 | 6 | 15 | 38 | |
| GSE47630 | ARID3C | 138715 | 1 | 20 | 19 | |
| GSE54993 | ARID3C | 138715 | 6 | 0 | 64 | |
| GSE54994 | ARID3C | 138715 | 6 | 12 | 35 | |
| GSE60625 | ARID3C | 138715 | 0 | 0 | 11 | |
| GSE74703 | ARID3C | 138715 | 2 | 7 | 27 | |
| GSE74704 | ARID3C | 138715 | 0 | 11 | 9 | |
| TCGA | ARID3C | 138715 | 17 | 44 | 35 |
Total number of gains: 49; Total number of losses: 145; Total Number of normals: 294.
Somatic mutations of ARID3C:
Generating mutation plots.
Highly correlated genes for ARID3C:
Showing all 8 correlated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARID3C | TH | 0.739396 | 3 | 0 | 3 |
| ARID3C | SLC4A1 | 0.688716 | 3 | 0 | 3 |
| ARID3C | PLA2G2F | 0.645842 | 3 | 0 | 3 |
| ARID3C | CDH22 | 0.604017 | 3 | 0 | 3 |
| ARID3C | PGLYRP2 | 0.589989 | 3 | 0 | 3 |
| ARID3C | SLC39A5 | 0.577084 | 3 | 0 | 3 |
| ARID3C | RIMBP2 | 0.556639 | 3 | 0 | 3 |
| ARID3C | GFRA4 | 0.551612 | 3 | 0 | 3 |
For details and further investigation, click here