| Full name: tyrosine hydroxylase | Alias Symbol: DYT5b | ||
| Type: protein-coding gene | Cytoband: 11p15.5 | ||
| Entrez ID: 7054 | HGNC ID: HGNC:11782 | Ensembl Gene: ENSG00000180176 | OMIM ID: 191290 |
| Drug and gene relationship at DGIdb | |||
TH involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04728 | Dopaminergic synapse | |
| hsa04917 | Prolactin signaling pathway | |
| hsa05031 | Amphetamine addiction |
Expression of TH:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TH | 7054 | 208291_s_at | -0.0215 | 0.9714 | |
| GSE20347 | TH | 7054 | 208291_s_at | -0.0770 | 0.1767 | |
| GSE23400 | TH | 7054 | 208291_s_at | -0.1000 | 0.0149 | |
| GSE26886 | TH | 7054 | 208291_s_at | -0.0280 | 0.8853 | |
| GSE29001 | TH | 7054 | 208291_s_at | 0.0231 | 0.9143 | |
| GSE38129 | TH | 7054 | 208291_s_at | -0.1102 | 0.0975 | |
| GSE45670 | TH | 7054 | 208291_s_at | 0.0649 | 0.7770 | |
| GSE53622 | TH | 7054 | 150816 | 0.0846 | 0.3652 | |
| GSE53624 | TH | 7054 | 20540 | -0.9206 | 0.0000 | |
| GSE63941 | TH | 7054 | 208291_s_at | 0.2289 | 0.1050 | |
| GSE77861 | TH | 7054 | 208291_s_at | -0.2324 | 0.0933 | |
| GSE97050 | TH | 7054 | A_33_P3252414 | 0.1931 | 0.3765 | |
| TCGA | TH | 7054 | RNAseq | -0.0881 | 0.9148 |
Upregulated datasets: 0; Downregulated datasets: 0.
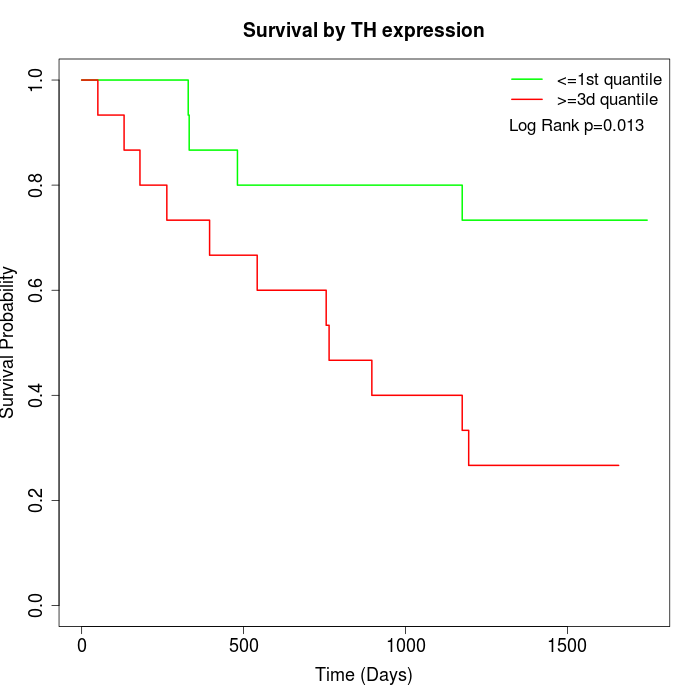
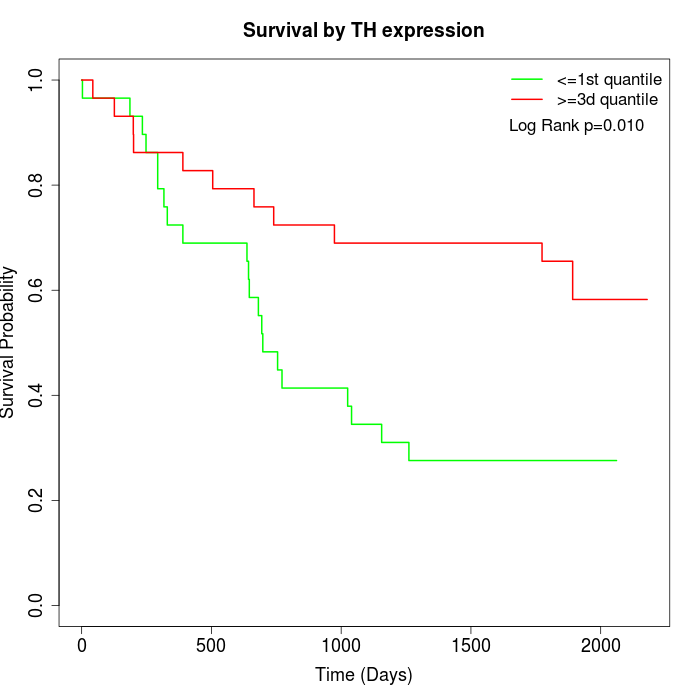
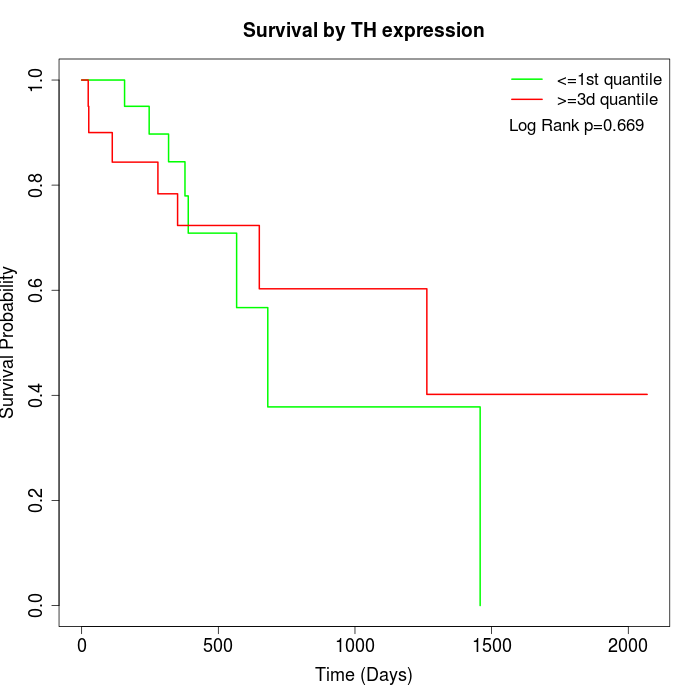
Survival by TH expression:
Note: Click image to view full size file.
Copy number change of TH:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TH | 7054 | 1 | 11 | 18 | |
| GSE20123 | TH | 7054 | 1 | 12 | 17 | |
| GSE43470 | TH | 7054 | 1 | 9 | 33 | |
| GSE46452 | TH | 7054 | 7 | 8 | 44 | |
| GSE47630 | TH | 7054 | 4 | 12 | 24 | |
| GSE54993 | TH | 7054 | 3 | 1 | 66 | |
| GSE54994 | TH | 7054 | 1 | 12 | 40 | |
| GSE60625 | TH | 7054 | 0 | 0 | 11 | |
| GSE74703 | TH | 7054 | 1 | 7 | 28 | |
| GSE74704 | TH | 7054 | 0 | 8 | 12 | |
| TCGA | TH | 7054 | 8 | 39 | 49 |
Total number of gains: 27; Total number of losses: 119; Total Number of normals: 342.
Somatic mutations of TH:
Generating mutation plots.
Highly correlated genes for TH:
Showing top 20/271 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TH | SUN5 | 0.769996 | 3 | 0 | 3 |
| TH | GPR152 | 0.75224 | 3 | 0 | 3 |
| TH | ARID3C | 0.739396 | 3 | 0 | 3 |
| TH | SLC39A5 | 0.725635 | 3 | 0 | 3 |
| TH | SLC22A31 | 0.72152 | 5 | 0 | 5 |
| TH | OR1J4 | 0.717679 | 3 | 0 | 3 |
| TH | PNPLA7 | 0.709607 | 3 | 0 | 3 |
| TH | DEGS2 | 0.703611 | 3 | 0 | 3 |
| TH | SLC24A4 | 0.698953 | 3 | 0 | 3 |
| TH | PTAFR | 0.679367 | 3 | 0 | 3 |
| TH | TOR2A | 0.679224 | 4 | 0 | 4 |
| TH | FAM222A-AS1 | 0.677197 | 3 | 0 | 3 |
| TH | GABRB3 | 0.676081 | 3 | 0 | 3 |
| TH | C14orf180 | 0.666918 | 3 | 0 | 3 |
| TH | CLRN2 | 0.664744 | 3 | 0 | 3 |
| TH | S100A9 | 0.663588 | 3 | 0 | 3 |
| TH | MIATNB | 0.663377 | 3 | 0 | 3 |
| TH | GLTPD2 | 0.663012 | 3 | 0 | 3 |
| TH | GJD4 | 0.659744 | 4 | 0 | 4 |
| TH | RETN | 0.658549 | 4 | 0 | 4 |
For details and further investigation, click here