| Full name: peptidoglycan recognition protein 2 | Alias Symbol: PGRP-L|PGLYRPL|TAGL-like|tagL|tagL-alpha|tagl-beta|PGRPL | ||
| Type: protein-coding gene | Cytoband: 19p13.12 | ||
| Entrez ID: 114770 | HGNC ID: HGNC:30013 | Ensembl Gene: ENSG00000161031 | OMIM ID: 608199 |
| Drug and gene relationship at DGIdb | |||
Expression of PGLYRP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PGLYRP2 | 114770 | 242817_at | -0.0499 | 0.8655 | |
| GSE26886 | PGLYRP2 | 114770 | 242817_at | 0.2231 | 0.0700 | |
| GSE45670 | PGLYRP2 | 114770 | 242817_at | 0.0539 | 0.6408 | |
| GSE53622 | PGLYRP2 | 114770 | 4837 | -0.0747 | 0.3118 | |
| GSE53624 | PGLYRP2 | 114770 | 4837 | -0.0452 | 0.6028 | |
| GSE63941 | PGLYRP2 | 114770 | 242817_at | -0.0244 | 0.8973 | |
| GSE77861 | PGLYRP2 | 114770 | 242817_at | -0.0325 | 0.8734 | |
| GSE97050 | PGLYRP2 | 114770 | A_33_P3259917 | -0.1525 | 0.4854 | |
| TCGA | PGLYRP2 | 114770 | RNAseq | 1.7226 | 0.0431 |
Upregulated datasets: 1; Downregulated datasets: 0.
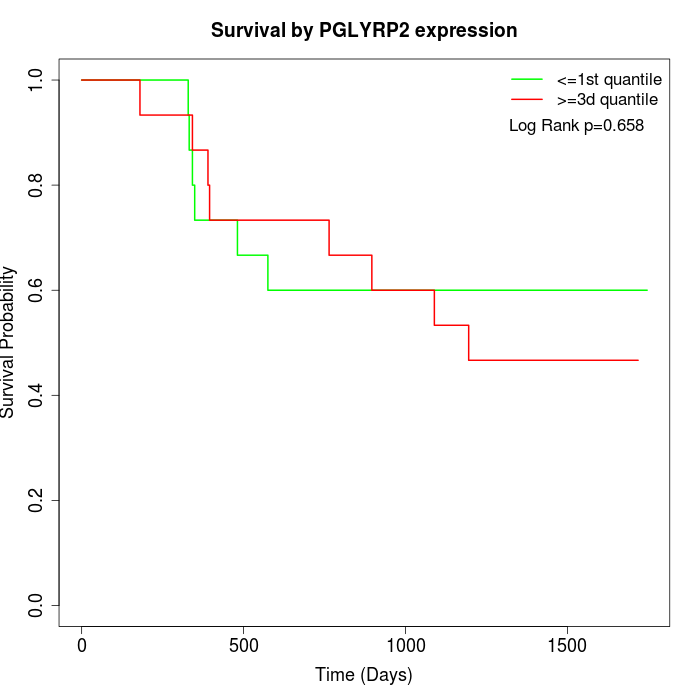
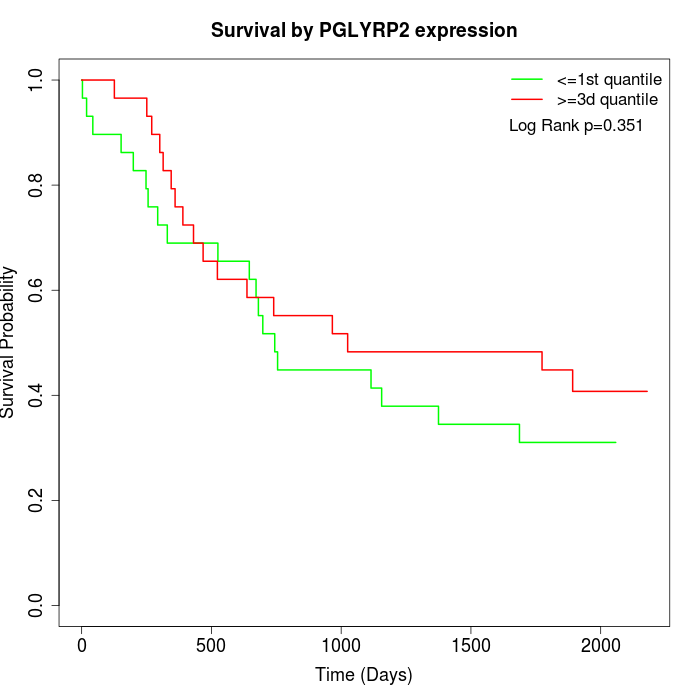
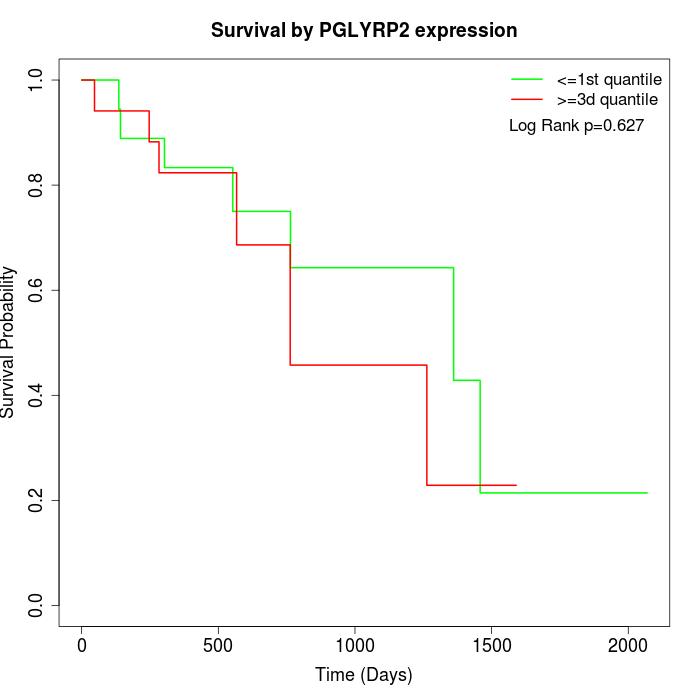
Survival by PGLYRP2 expression:
Note: Click image to view full size file.
Copy number change of PGLYRP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PGLYRP2 | 114770 | 4 | 2 | 24 | |
| GSE20123 | PGLYRP2 | 114770 | 3 | 1 | 26 | |
| GSE43470 | PGLYRP2 | 114770 | 2 | 6 | 35 | |
| GSE46452 | PGLYRP2 | 114770 | 47 | 1 | 11 | |
| GSE47630 | PGLYRP2 | 114770 | 4 | 7 | 29 | |
| GSE54993 | PGLYRP2 | 114770 | 15 | 4 | 51 | |
| GSE54994 | PGLYRP2 | 114770 | 6 | 14 | 33 | |
| GSE60625 | PGLYRP2 | 114770 | 9 | 0 | 2 | |
| GSE74703 | PGLYRP2 | 114770 | 2 | 4 | 30 | |
| GSE74704 | PGLYRP2 | 114770 | 0 | 1 | 19 | |
| TCGA | PGLYRP2 | 114770 | 20 | 10 | 66 |
Total number of gains: 112; Total number of losses: 50; Total Number of normals: 326.
Somatic mutations of PGLYRP2:
Generating mutation plots.
Highly correlated genes for PGLYRP2:
Showing top 20/180 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PGLYRP2 | SPACA3 | 0.747673 | 4 | 0 | 4 |
| PGLYRP2 | ANKDD1A | 0.745793 | 3 | 0 | 3 |
| PGLYRP2 | CDH22 | 0.743944 | 3 | 0 | 3 |
| PGLYRP2 | C19orf25 | 0.73336 | 3 | 0 | 3 |
| PGLYRP2 | LHX9 | 0.719325 | 5 | 0 | 4 |
| PGLYRP2 | FGF17 | 0.71699 | 3 | 0 | 3 |
| PGLYRP2 | ROM1 | 0.715994 | 3 | 0 | 3 |
| PGLYRP2 | PRAP1 | 0.712781 | 3 | 0 | 3 |
| PGLYRP2 | CCL25 | 0.711254 | 4 | 0 | 3 |
| PGLYRP2 | FGF22 | 0.708403 | 3 | 0 | 3 |
| PGLYRP2 | STRC | 0.698285 | 3 | 0 | 3 |
| PGLYRP2 | REG3A | 0.696863 | 3 | 0 | 3 |
| PGLYRP2 | C2orf50 | 0.696327 | 3 | 0 | 3 |
| PGLYRP2 | PGLYRP1 | 0.694111 | 4 | 0 | 3 |
| PGLYRP2 | AGER | 0.693319 | 3 | 0 | 3 |
| PGLYRP2 | C2CD4D | 0.686302 | 3 | 0 | 3 |
| PGLYRP2 | ADAM33 | 0.685833 | 3 | 0 | 3 |
| PGLYRP2 | LTC4S | 0.683755 | 4 | 0 | 3 |
| PGLYRP2 | NPPA | 0.683267 | 3 | 0 | 3 |
| PGLYRP2 | TTC34 | 0.681699 | 3 | 0 | 3 |
For details and further investigation, click here