| Full name: cancer susceptibility 11 | Alias Symbol: TCONS_00014535|LINC00990|CARLo-7|ENST00000518376 | ||
| Type: non-coding RNA | Cytoband: 8q24.21 | ||
| Entrez ID: 100270680 | HGNC ID: HGNC:48939 | Ensembl Gene: ENSG00000249375 | OMIM ID: 617704 |
| Drug and gene relationship at DGIdb | |||
Expression of CASC11:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CASC11 | 100270680 | 1561232_at | 0.0297 | 0.9206 | |
| GSE26886 | CASC11 | 100270680 | 1561232_at | 0.0275 | 0.8012 | |
| GSE45670 | CASC11 | 100270680 | 1561232_at | 0.0210 | 0.8123 | |
| GSE53622 | CASC11 | 100270680 | 42545 | 0.4304 | 0.0004 | |
| GSE53624 | CASC11 | 100270680 | 140824 | 0.5648 | 0.0000 | |
| GSE63941 | CASC11 | 100270680 | 1561232_at | 0.0413 | 0.8166 | |
| GSE77861 | CASC11 | 100270680 | 1561232_at | -0.0342 | 0.7192 | |
| SRP133303 | CASC11 | 100270680 | RNAseq | 0.1026 | 0.6131 |
Upregulated datasets: 0; Downregulated datasets: 0.
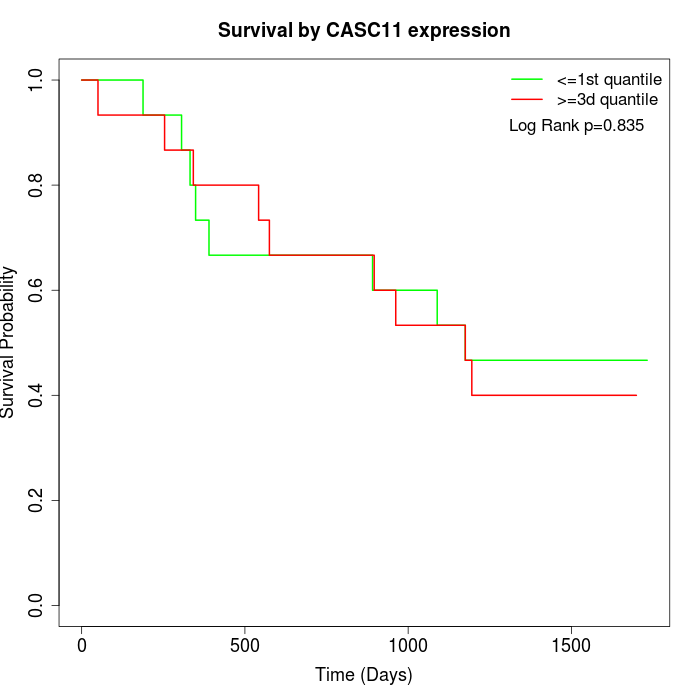
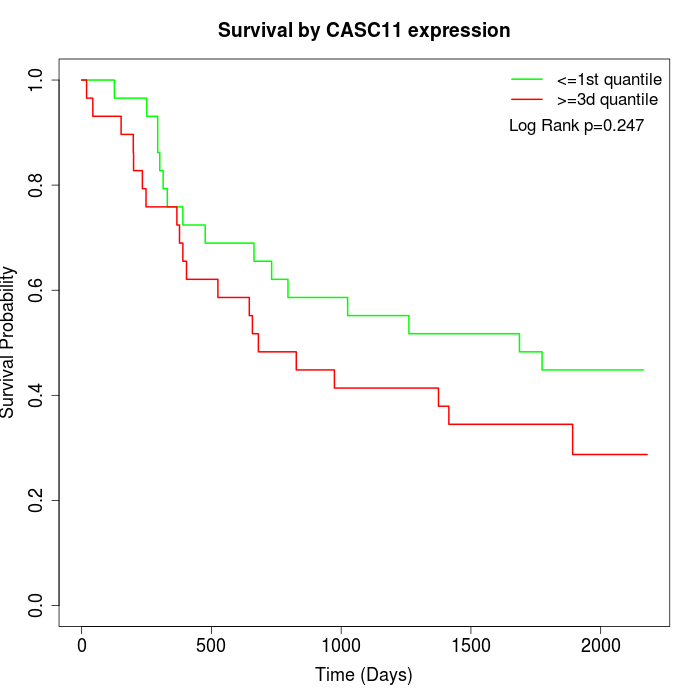
Survival by CASC11 expression:
Note: Click image to view full size file.
Copy number change of CASC11:
No record found for this gene.
Somatic mutations of CASC11:
Generating mutation plots.
Highly correlated genes for CASC11:
Showing all 17 correlated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CASC11 | CNGB3 | 0.642565 | 3 | 0 | 3 |
| CASC11 | SRRM4 | 0.639838 | 3 | 0 | 3 |
| CASC11 | LONRF3 | 0.635803 | 3 | 0 | 3 |
| CASC11 | CCDC140 | 0.630522 | 3 | 0 | 3 |
| CASC11 | NXPE1 | 0.599155 | 3 | 0 | 3 |
| CASC11 | TMEM26 | 0.598646 | 4 | 0 | 3 |
| CASC11 | OR10H3 | 0.587695 | 3 | 0 | 3 |
| CASC11 | GATA3-AS1 | 0.58219 | 4 | 0 | 3 |
| CASC11 | TAL2 | 0.581809 | 4 | 0 | 3 |
| CASC11 | DPYSL5 | 0.566049 | 4 | 0 | 3 |
| CASC11 | ACBD4 | 0.545246 | 3 | 0 | 3 |
| CASC11 | LINC01007 | 0.542942 | 4 | 0 | 3 |
| CASC11 | CELF2-AS1 | 0.540136 | 4 | 0 | 3 |
| CASC11 | ARMCX4 | 0.535564 | 3 | 0 | 3 |
| CASC11 | TNFSF11 | 0.521878 | 4 | 0 | 3 |
| CASC11 | OR5V1 | 0.511974 | 3 | 0 | 3 |
| CASC11 | ARHGEF7-IT1 | 0.507573 | 4 | 0 | 3 |
For details and further investigation, click here