| Full name: DDB1 and CUL4 associated factor 4 like 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 8q21.3 | ||
| Entrez ID: 138009 | HGNC ID: HGNC:26657 | Ensembl Gene: ENSG00000176566 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of DCAF4L2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DCAF4L2 | 138009 | 1553197_at | -0.1247 | 0.5485 | |
| GSE26886 | DCAF4L2 | 138009 | 1553197_at | 0.0376 | 0.7153 | |
| GSE45670 | DCAF4L2 | 138009 | 1553197_at | 0.1719 | 0.6001 | |
| GSE53622 | DCAF4L2 | 138009 | 26106 | 0.8704 | 0.0000 | |
| GSE53624 | DCAF4L2 | 138009 | 26106 | 0.3184 | 0.0361 | |
| GSE63941 | DCAF4L2 | 138009 | 1553197_at | -0.0663 | 0.6077 | |
| GSE77861 | DCAF4L2 | 138009 | 1553199_at | -0.0278 | 0.8374 | |
| TCGA | DCAF4L2 | 138009 | RNAseq | 3.4793 | 0.0756 |
Upregulated datasets: 0; Downregulated datasets: 0.
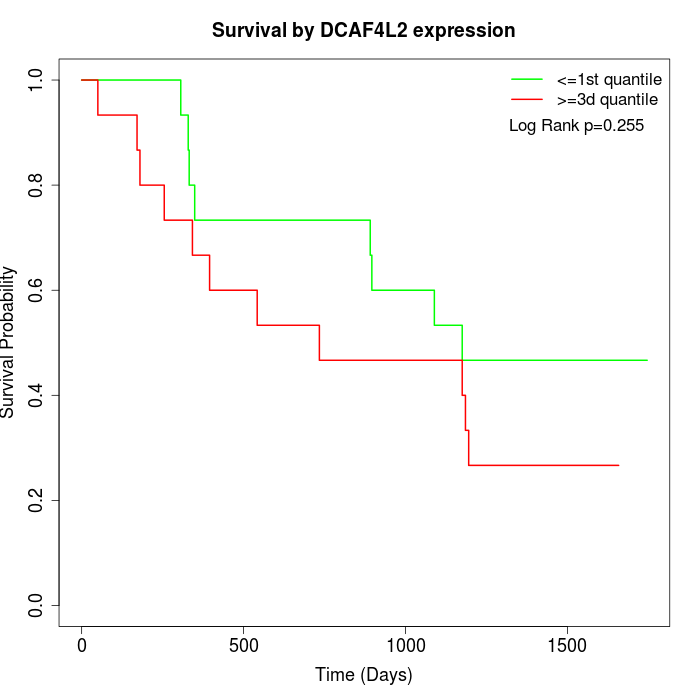
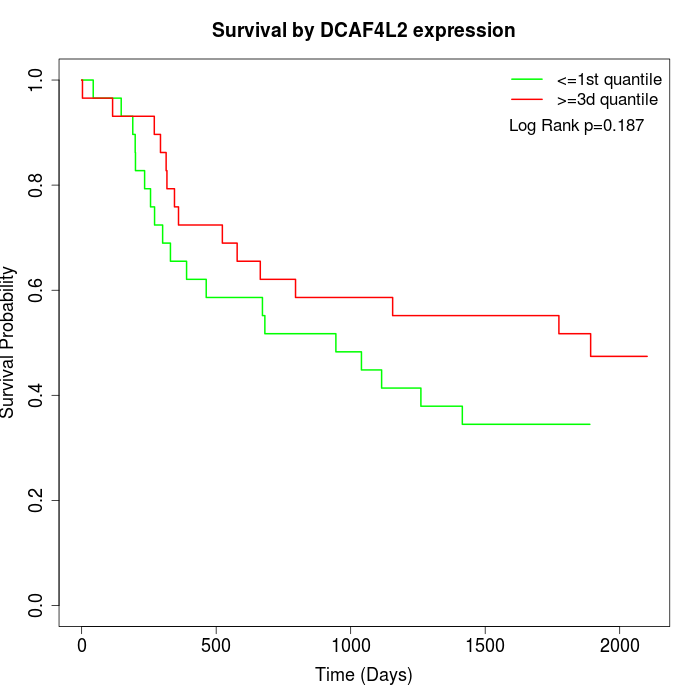
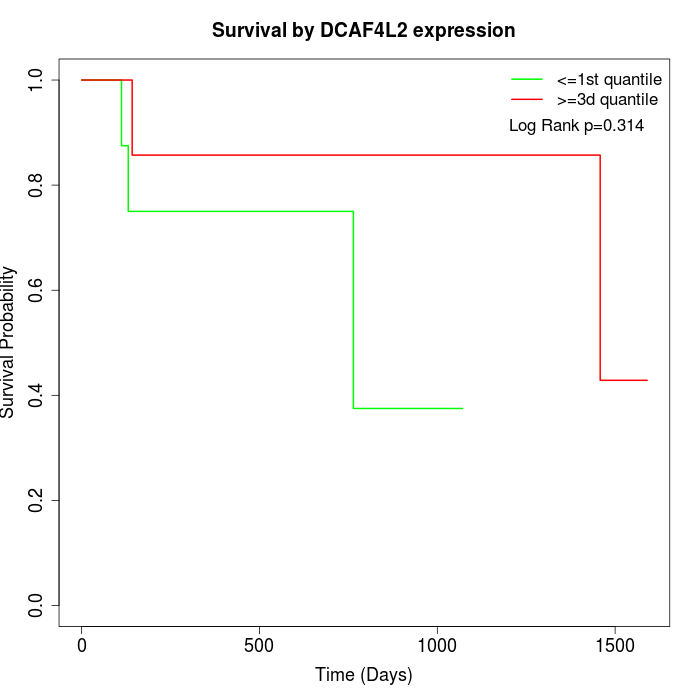
Survival by DCAF4L2 expression:
Note: Click image to view full size file.
Copy number change of DCAF4L2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DCAF4L2 | 138009 | 14 | 1 | 15 | |
| GSE20123 | DCAF4L2 | 138009 | 16 | 1 | 13 | |
| GSE43470 | DCAF4L2 | 138009 | 19 | 1 | 23 | |
| GSE46452 | DCAF4L2 | 138009 | 21 | 0 | 38 | |
| GSE47630 | DCAF4L2 | 138009 | 23 | 0 | 17 | |
| GSE54993 | DCAF4L2 | 138009 | 0 | 20 | 50 | |
| GSE54994 | DCAF4L2 | 138009 | 33 | 1 | 19 | |
| GSE60625 | DCAF4L2 | 138009 | 0 | 4 | 7 | |
| GSE74703 | DCAF4L2 | 138009 | 16 | 1 | 19 | |
| GSE74704 | DCAF4L2 | 138009 | 10 | 0 | 10 | |
| TCGA | DCAF4L2 | 138009 | 52 | 3 | 41 |
Total number of gains: 204; Total number of losses: 32; Total Number of normals: 252.
Somatic mutations of DCAF4L2:
Generating mutation plots.
Highly correlated genes for DCAF4L2:
Showing top 20/34 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DCAF4L2 | GRIA4 | 0.748204 | 3 | 0 | 3 |
| DCAF4L2 | LINC00276 | 0.711568 | 3 | 0 | 3 |
| DCAF4L2 | LINC01350 | 0.696763 | 3 | 0 | 3 |
| DCAF4L2 | C8orf74 | 0.682356 | 3 | 0 | 3 |
| DCAF4L2 | TEX35 | 0.669261 | 4 | 0 | 4 |
| DCAF4L2 | RAX2 | 0.668956 | 3 | 0 | 3 |
| DCAF4L2 | ZNF248 | 0.663463 | 3 | 0 | 3 |
| DCAF4L2 | MAGEB1 | 0.644635 | 5 | 0 | 4 |
| DCAF4L2 | OXTR | 0.63853 | 3 | 0 | 3 |
| DCAF4L2 | DEFB108B | 0.636213 | 3 | 0 | 3 |
| DCAF4L2 | SLC26A1 | 0.631456 | 3 | 0 | 3 |
| DCAF4L2 | SLC22A7 | 0.618342 | 4 | 0 | 3 |
| DCAF4L2 | LINC01366 | 0.611053 | 3 | 0 | 3 |
| DCAF4L2 | FAM71F1 | 0.609301 | 3 | 0 | 3 |
| DCAF4L2 | TRAPPC3L | 0.598763 | 4 | 0 | 3 |
| DCAF4L2 | ASB15 | 0.593664 | 3 | 0 | 3 |
| DCAF4L2 | ELOVL3 | 0.59308 | 3 | 0 | 3 |
| DCAF4L2 | FAM222A-AS1 | 0.586454 | 3 | 0 | 3 |
| DCAF4L2 | VWA5B1 | 0.582853 | 4 | 0 | 3 |
| DCAF4L2 | CTAG2 | 0.578121 | 4 | 0 | 3 |
For details and further investigation, click here