| Full name: retina and anterior neural fold homeobox 2 | Alias Symbol: MGC15631|ARMD6|CORD11 | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 84839 | HGNC ID: HGNC:18286 | Ensembl Gene: ENSG00000173976 | OMIM ID: 610362 |
| Drug and gene relationship at DGIdb | |||
Expression of RAX2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RAX2 | 84839 | 1552311_a_at | -0.0629 | 0.7951 | |
| GSE26886 | RAX2 | 84839 | 1552311_a_at | 0.0810 | 0.5770 | |
| GSE45670 | RAX2 | 84839 | 1552311_a_at | 0.1037 | 0.1558 | |
| GSE53622 | RAX2 | 84839 | 29449 | 0.5203 | 0.0000 | |
| GSE53624 | RAX2 | 84839 | 29449 | 0.1813 | 0.0528 | |
| GSE63941 | RAX2 | 84839 | 1552311_a_at | 0.1598 | 0.2200 | |
| GSE77861 | RAX2 | 84839 | 1552311_a_at | -0.0581 | 0.6854 | |
| GSE97050 | RAX2 | 84839 | A_23_P56146 | -0.0457 | 0.8864 | |
| SRP133303 | RAX2 | 84839 | RNAseq | -0.4343 | 0.0172 | |
| SRP219564 | RAX2 | 84839 | RNAseq | 0.4398 | 0.4722 |
Upregulated datasets: 0; Downregulated datasets: 0.
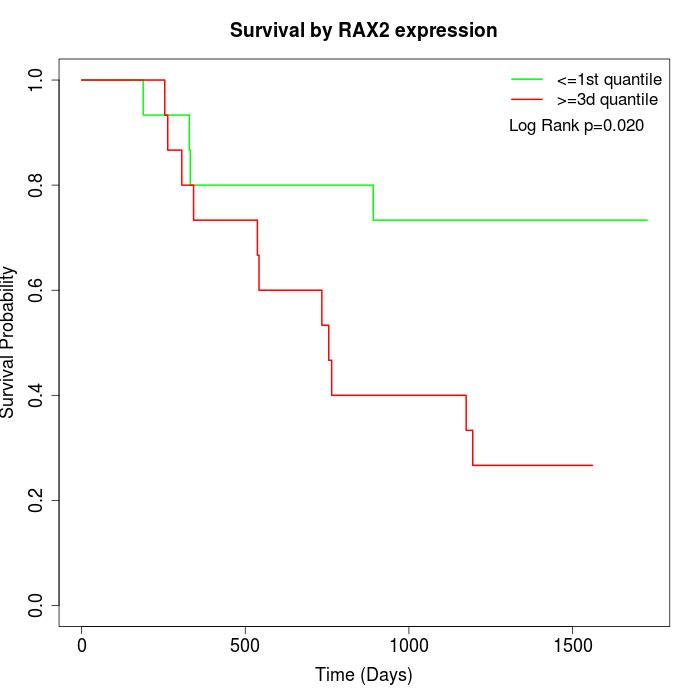
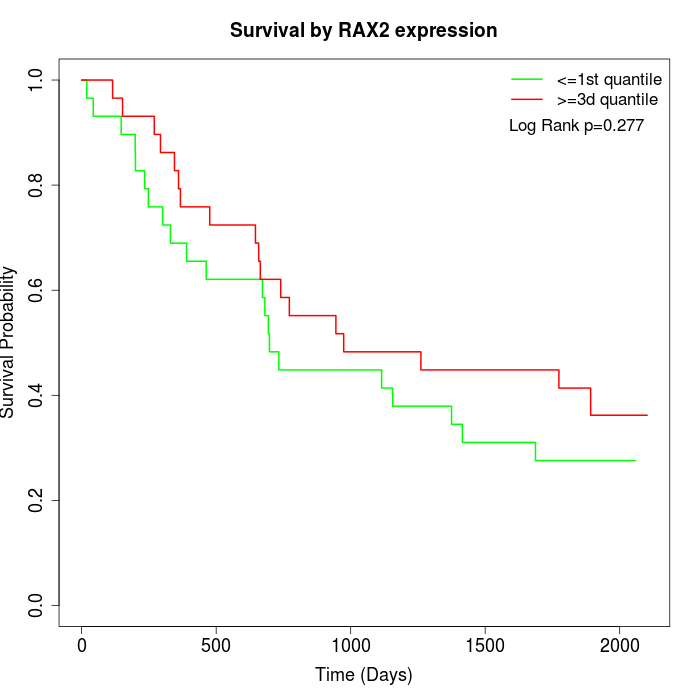
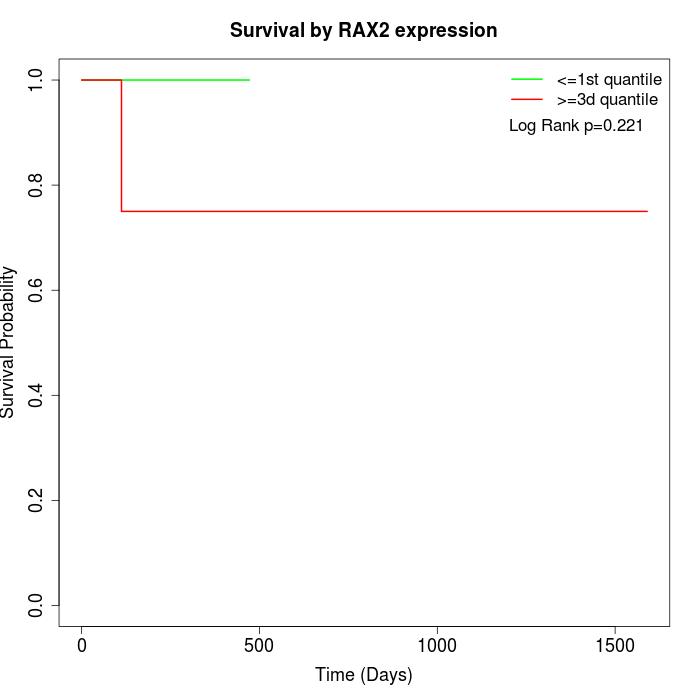
Survival by RAX2 expression:
Note: Click image to view full size file.
Copy number change of RAX2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RAX2 | 84839 | 5 | 3 | 22 | |
| GSE20123 | RAX2 | 84839 | 4 | 2 | 24 | |
| GSE43470 | RAX2 | 84839 | 1 | 9 | 33 | |
| GSE46452 | RAX2 | 84839 | 47 | 1 | 11 | |
| GSE47630 | RAX2 | 84839 | 5 | 7 | 28 | |
| GSE54993 | RAX2 | 84839 | 16 | 3 | 51 | |
| GSE54994 | RAX2 | 84839 | 8 | 16 | 29 | |
| GSE60625 | RAX2 | 84839 | 9 | 0 | 2 | |
| GSE74703 | RAX2 | 84839 | 1 | 6 | 29 | |
| GSE74704 | RAX2 | 84839 | 1 | 2 | 17 | |
| TCGA | RAX2 | 84839 | 10 | 19 | 67 |
Total number of gains: 107; Total number of losses: 68; Total Number of normals: 313.
Somatic mutations of RAX2:
Generating mutation plots.
Highly correlated genes for RAX2:
Showing top 20/422 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RAX2 | ZNF527 | 0.837981 | 4 | 0 | 4 |
| RAX2 | BMP8B | 0.815507 | 4 | 0 | 4 |
| RAX2 | DCDC1 | 0.808059 | 3 | 0 | 3 |
| RAX2 | SLC6A3 | 0.807837 | 3 | 0 | 3 |
| RAX2 | LBX2 | 0.804437 | 3 | 0 | 3 |
| RAX2 | FOXE3 | 0.7884 | 3 | 0 | 3 |
| RAX2 | YY2 | 0.784479 | 3 | 0 | 3 |
| RAX2 | RBM28 | 0.778053 | 3 | 0 | 3 |
| RAX2 | STRC | 0.773733 | 3 | 0 | 3 |
| RAX2 | CHST10 | 0.770796 | 4 | 0 | 4 |
| RAX2 | LINC00276 | 0.769497 | 3 | 0 | 3 |
| RAX2 | GLI4 | 0.76373 | 3 | 0 | 3 |
| RAX2 | KRTAP10-12 | 0.760861 | 3 | 0 | 3 |
| RAX2 | NTNG2 | 0.754421 | 4 | 0 | 4 |
| RAX2 | VWA5B1 | 0.753035 | 3 | 0 | 3 |
| RAX2 | KANK3 | 0.752761 | 3 | 0 | 3 |
| RAX2 | FAM30A | 0.745656 | 3 | 0 | 3 |
| RAX2 | C9orf153 | 0.737728 | 3 | 0 | 3 |
| RAX2 | GHSR | 0.73766 | 3 | 0 | 3 |
| RAX2 | F2RL3 | 0.7352 | 5 | 0 | 5 |
For details and further investigation, click here