| Full name: MIR31 host gene | Alias Symbol: LOC554202|hsa-lnc-31|LncHIFCAR | ||
| Type: non-coding RNA | Cytoband: 9p21.3 | ||
| Entrez ID: 554202 | HGNC ID: HGNC:37187 | Ensembl Gene: ENSG00000171889 | OMIM ID: 616356 |
| Drug and gene relationship at DGIdb | |||
Expression of MIR31HG:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MIR31HG | 554202 | 1554097_a_at | -0.4165 | 0.6995 | |
| GSE26886 | MIR31HG | 554202 | 1554097_a_at | 0.2167 | 0.6624 | |
| GSE45670 | MIR31HG | 554202 | 1554097_a_at | 1.2889 | 0.0181 | |
| GSE53622 | MIR31HG | 554202 | 48472 | 0.6469 | 0.0225 | |
| GSE53624 | MIR31HG | 554202 | 48472 | 0.1644 | 0.4699 | |
| GSE63941 | MIR31HG | 554202 | 1554097_a_at | -1.7587 | 0.1245 | |
| GSE77861 | MIR31HG | 554202 | 1554097_a_at | 0.1543 | 0.7462 | |
| SRP007169 | MIR31HG | 554202 | RNAseq | 0.0581 | 0.9342 | |
| SRP133303 | MIR31HG | 554202 | RNAseq | 1.6880 | 0.0122 | |
| SRP159526 | MIR31HG | 554202 | RNAseq | -0.4998 | 0.5523 |
Upregulated datasets: 2; Downregulated datasets: 0.
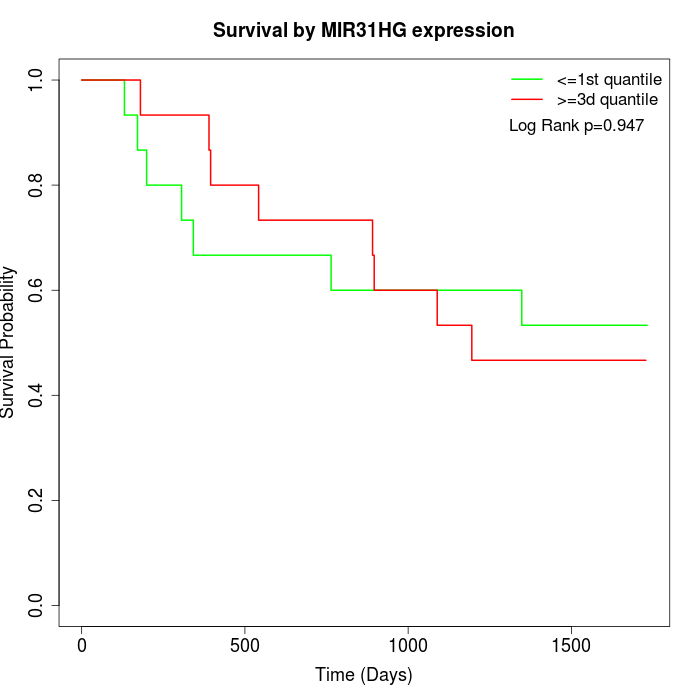
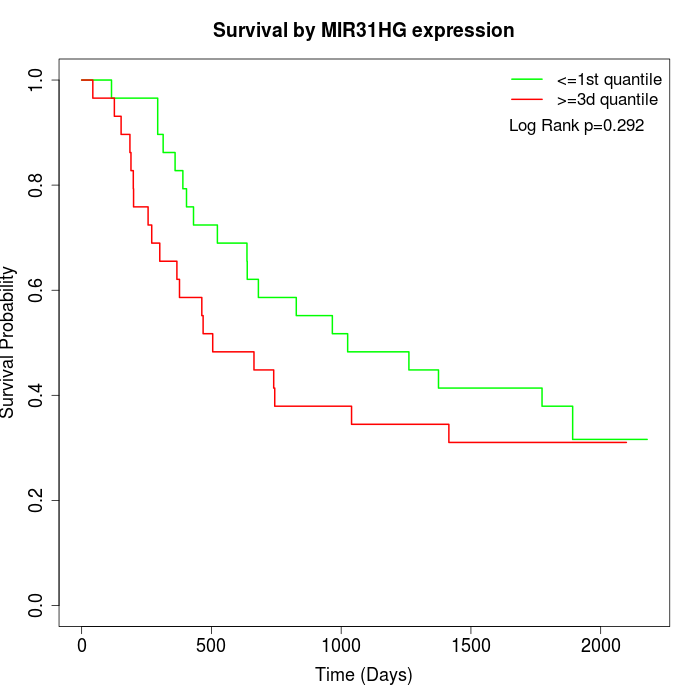
Survival by MIR31HG expression:
Note: Click image to view full size file.
Copy number change of MIR31HG:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MIR31HG | 554202 | 2 | 17 | 11 | |
| GSE20123 | MIR31HG | 554202 | 2 | 18 | 10 | |
| GSE43470 | MIR31HG | 554202 | 3 | 13 | 27 | |
| GSE46452 | MIR31HG | 554202 | 1 | 26 | 32 | |
| GSE47630 | MIR31HG | 554202 | 3 | 27 | 10 | |
| GSE54993 | MIR31HG | 554202 | 8 | 2 | 60 | |
| GSE54994 | MIR31HG | 554202 | 7 | 18 | 28 | |
| GSE60625 | MIR31HG | 554202 | 3 | 0 | 8 | |
| GSE74703 | MIR31HG | 554202 | 3 | 10 | 23 | |
| GSE74704 | MIR31HG | 554202 | 0 | 13 | 7 | |
| TCGA | MIR31HG | 554202 | 11 | 55 | 30 |
Total number of gains: 43; Total number of losses: 199; Total Number of normals: 246.
Somatic mutations of MIR31HG:
Generating mutation plots.
Highly correlated genes for MIR31HG:
Showing all 14 correlated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MIR31HG | IFNE | 0.786473 | 7 | 0 | 6 |
| MIR31HG | GXYLT2 | 0.655442 | 3 | 0 | 3 |
| MIR31HG | UBAC2-AS1 | 0.652439 | 3 | 0 | 3 |
| MIR31HG | MEG3 | 0.580656 | 3 | 0 | 3 |
| MIR31HG | RGS20 | 0.574156 | 4 | 0 | 4 |
| MIR31HG | NDRG1 | 0.567618 | 4 | 0 | 3 |
| MIR31HG | TRIM32 | 0.556816 | 4 | 0 | 4 |
| MIR31HG | NCAPG | 0.555227 | 3 | 0 | 3 |
| MIR31HG | TRIM5 | 0.54469 | 4 | 0 | 3 |
| MIR31HG | ZP4 | 0.534528 | 3 | 0 | 3 |
| MIR31HG | LINC01094 | 0.528248 | 4 | 0 | 3 |
| MIR31HG | HMMR-AS1 | 0.521945 | 4 | 0 | 3 |
| MIR31HG | LINC01137 | 0.515164 | 6 | 0 | 3 |
| MIR31HG | AGPAT4 | 0.512224 | 4 | 0 | 3 |
For details and further investigation, click here