| Full name: protein tyrosine phosphatase receptor type M | Alias Symbol: RPTPU|hR-PTPu | ||
| Type: protein-coding gene | Cytoband: 18p11.23 | ||
| Entrez ID: 5797 | HGNC ID: HGNC:9675 | Ensembl Gene: ENSG00000173482 | OMIM ID: 176888 |
| Drug and gene relationship at DGIdb | |||
PTPRM involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04520 | Adherens junction |
Expression of PTPRM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PTPRM | 5797 | 1555579_s_at | -0.6251 | 0.3774 | |
| GSE20347 | PTPRM | 5797 | 203329_at | 0.4537 | 0.0033 | |
| GSE23400 | PTPRM | 5797 | 203329_at | 0.1829 | 0.0311 | |
| GSE26886 | PTPRM | 5797 | 1555579_s_at | 1.6347 | 0.0000 | |
| GSE29001 | PTPRM | 5797 | 203329_at | 0.4783 | 0.1253 | |
| GSE38129 | PTPRM | 5797 | 203329_at | -0.0809 | 0.7895 | |
| GSE45670 | PTPRM | 5797 | 1555579_s_at | -0.9742 | 0.0184 | |
| GSE53622 | PTPRM | 5797 | 55356 | -0.1361 | 0.2716 | |
| GSE53624 | PTPRM | 5797 | 55356 | 0.2055 | 0.1061 | |
| GSE63941 | PTPRM | 5797 | 203329_at | -2.6746 | 0.0141 | |
| GSE77861 | PTPRM | 5797 | 1555579_s_at | 0.5045 | 0.0343 | |
| GSE97050 | PTPRM | 5797 | A_23_P159255 | -0.0903 | 0.7079 | |
| SRP007169 | PTPRM | 5797 | RNAseq | 2.0611 | 0.0000 | |
| SRP008496 | PTPRM | 5797 | RNAseq | 2.9400 | 0.0000 | |
| SRP064894 | PTPRM | 5797 | RNAseq | 0.5717 | 0.0916 | |
| SRP133303 | PTPRM | 5797 | RNAseq | 0.0868 | 0.7071 | |
| SRP159526 | PTPRM | 5797 | RNAseq | 1.0007 | 0.0290 | |
| SRP193095 | PTPRM | 5797 | RNAseq | 1.2467 | 0.0000 | |
| SRP219564 | PTPRM | 5797 | RNAseq | 0.4102 | 0.4815 | |
| TCGA | PTPRM | 5797 | RNAseq | -0.5565 | 0.0000 |
Upregulated datasets: 5; Downregulated datasets: 1.
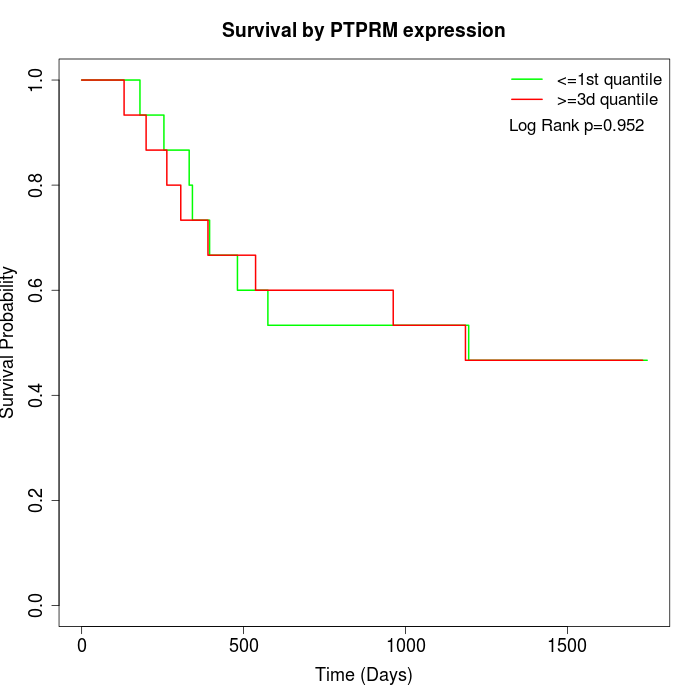
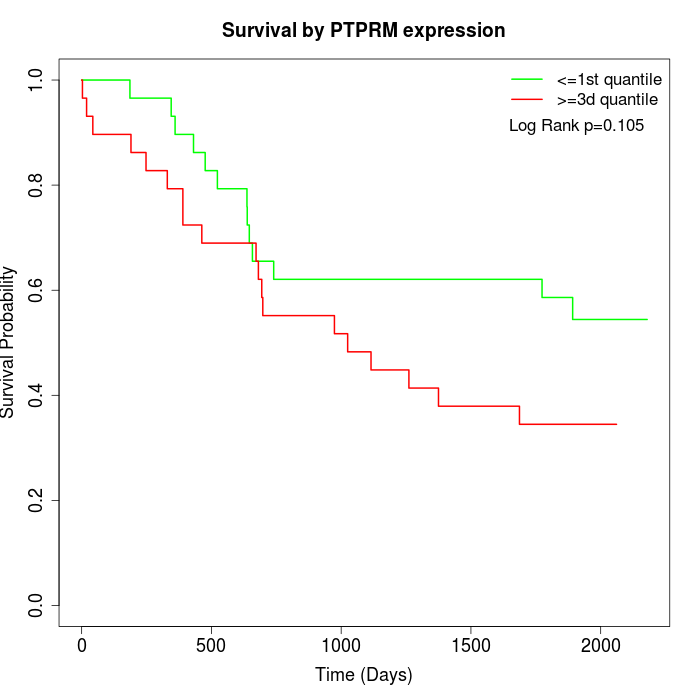
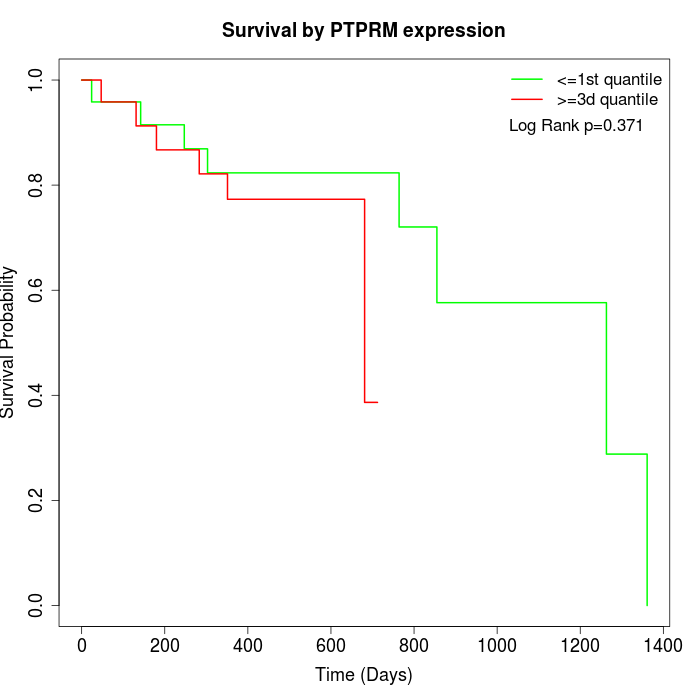
Survival by PTPRM expression:
Note: Click image to view full size file.
Copy number change of PTPRM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PTPRM | 5797 | 8 | 3 | 19 | |
| GSE20123 | PTPRM | 5797 | 8 | 3 | 19 | |
| GSE43470 | PTPRM | 5797 | 4 | 5 | 34 | |
| GSE46452 | PTPRM | 5797 | 3 | 21 | 35 | |
| GSE47630 | PTPRM | 5797 | 7 | 17 | 16 | |
| GSE54993 | PTPRM | 5797 | 6 | 3 | 61 | |
| GSE54994 | PTPRM | 5797 | 9 | 8 | 36 | |
| GSE60625 | PTPRM | 5797 | 0 | 4 | 7 | |
| GSE74703 | PTPRM | 5797 | 3 | 3 | 30 | |
| GSE74704 | PTPRM | 5797 | 5 | 2 | 13 | |
| TCGA | PTPRM | 5797 | 31 | 23 | 42 |
Total number of gains: 84; Total number of losses: 92; Total Number of normals: 312.
Somatic mutations of PTPRM:
Generating mutation plots.
Highly correlated genes for PTPRM:
Showing top 20/1339 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PTPRM | CYP4V2 | 0.829434 | 3 | 0 | 3 |
| PTPRM | RHOJ | 0.806988 | 4 | 0 | 4 |
| PTPRM | TP53I13 | 0.791842 | 3 | 0 | 3 |
| PTPRM | C11orf96 | 0.777159 | 5 | 0 | 5 |
| PTPRM | HAGH | 0.764195 | 3 | 0 | 3 |
| PTPRM | ZBTB26 | 0.751523 | 3 | 0 | 3 |
| PTPRM | WDR81 | 0.750305 | 3 | 0 | 3 |
| PTPRM | HSPA5 | 0.74328 | 3 | 0 | 3 |
| PTPRM | ANXA6 | 0.740914 | 12 | 0 | 11 |
| PTPRM | PDK2 | 0.738696 | 3 | 0 | 3 |
| PTPRM | PMP22 | 0.7379 | 12 | 0 | 10 |
| PTPRM | CLMP | 0.735769 | 6 | 0 | 6 |
| PTPRM | MYADM | 0.734406 | 6 | 0 | 5 |
| PTPRM | ATP10A | 0.727852 | 6 | 0 | 6 |
| PTPRM | FKBP7 | 0.723892 | 7 | 0 | 7 |
| PTPRM | DIXDC1 | 0.722961 | 8 | 0 | 8 |
| PTPRM | LAMA4 | 0.721183 | 12 | 0 | 12 |
| PTPRM | B3GNT9 | 0.719387 | 3 | 0 | 3 |
| PTPRM | GLIS2 | 0.717794 | 7 | 0 | 7 |
| PTPRM | CRISPLD2 | 0.716881 | 13 | 0 | 12 |
For details and further investigation, click here