| Full name: neuronal PAS domain protein 2 | Alias Symbol: MOP4|PASD4|bHLHe9 | ||
| Type: protein-coding gene | Cytoband: 2q11.2 | ||
| Entrez ID: 4862 | HGNC ID: HGNC:7895 | Ensembl Gene: ENSG00000170485 | OMIM ID: 603347 |
| Drug and gene relationship at DGIdb | |||
NPAS2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04710 | Circadian rhythm |
Expression of NPAS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NPAS2 | 4862 | 213462_at | -0.6888 | 0.5155 | |
| GSE20347 | NPAS2 | 4862 | 213462_at | -0.9969 | 0.0000 | |
| GSE23400 | NPAS2 | 4862 | 213462_at | -0.7771 | 0.0000 | |
| GSE26886 | NPAS2 | 4862 | 39549_at | -0.6980 | 0.0315 | |
| GSE29001 | NPAS2 | 4862 | 213462_at | -1.2381 | 0.0006 | |
| GSE38129 | NPAS2 | 4862 | 213462_at | -0.7673 | 0.0012 | |
| GSE45670 | NPAS2 | 4862 | 213462_at | -0.4246 | 0.0698 | |
| GSE63941 | NPAS2 | 4862 | 213462_at | -2.0170 | 0.0094 | |
| GSE77861 | NPAS2 | 4862 | 213462_at | -0.6914 | 0.0367 | |
| GSE97050 | NPAS2 | 4862 | A_23_P218597 | -0.2276 | 0.6132 | |
| SRP007169 | NPAS2 | 4862 | RNAseq | -3.0210 | 0.0000 | |
| SRP008496 | NPAS2 | 4862 | RNAseq | -2.5576 | 0.0000 | |
| SRP064894 | NPAS2 | 4862 | RNAseq | -1.4545 | 0.0000 | |
| SRP133303 | NPAS2 | 4862 | RNAseq | -1.3410 | 0.0000 | |
| SRP159526 | NPAS2 | 4862 | RNAseq | -0.7397 | 0.2884 | |
| SRP193095 | NPAS2 | 4862 | RNAseq | -1.2367 | 0.0000 | |
| SRP219564 | NPAS2 | 4862 | RNAseq | -1.2754 | 0.0956 | |
| TCGA | NPAS2 | 4862 | RNAseq | -0.2011 | 0.0523 |
Upregulated datasets: 0; Downregulated datasets: 7.
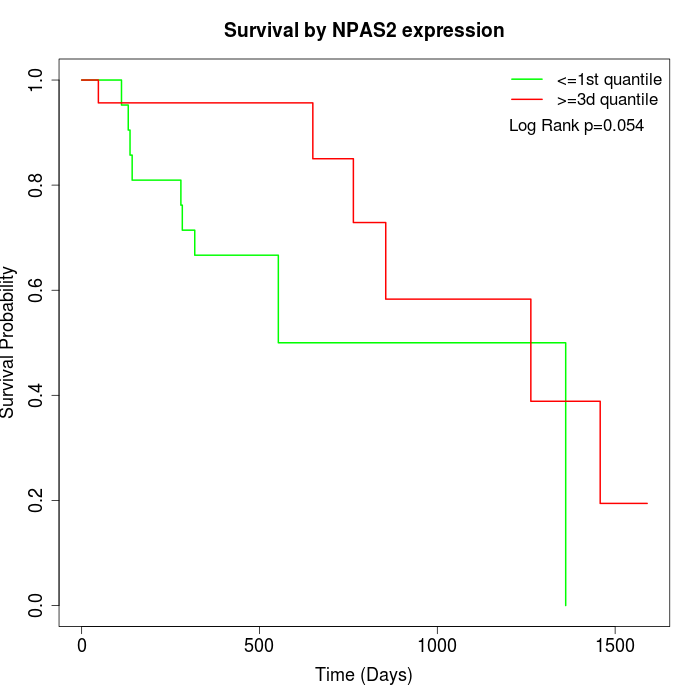
Survival by NPAS2 expression:
Note: Click image to view full size file.
Copy number change of NPAS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NPAS2 | 4862 | 5 | 2 | 23 | |
| GSE20123 | NPAS2 | 4862 | 5 | 2 | 23 | |
| GSE43470 | NPAS2 | 4862 | 5 | 1 | 37 | |
| GSE46452 | NPAS2 | 4862 | 1 | 4 | 54 | |
| GSE47630 | NPAS2 | 4862 | 7 | 0 | 33 | |
| GSE54993 | NPAS2 | 4862 | 0 | 6 | 64 | |
| GSE54994 | NPAS2 | 4862 | 10 | 0 | 43 | |
| GSE60625 | NPAS2 | 4862 | 0 | 3 | 8 | |
| GSE74703 | NPAS2 | 4862 | 5 | 1 | 30 | |
| GSE74704 | NPAS2 | 4862 | 3 | 1 | 16 | |
| TCGA | NPAS2 | 4862 | 34 | 5 | 57 |
Total number of gains: 75; Total number of losses: 25; Total Number of normals: 388.
Somatic mutations of NPAS2:
Generating mutation plots.
Highly correlated genes for NPAS2:
Showing top 20/1083 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NPAS2 | RNFT1 | 0.836834 | 3 | 0 | 3 |
| NPAS2 | TAB3 | 0.817058 | 4 | 0 | 4 |
| NPAS2 | HEATR5B | 0.767051 | 4 | 0 | 4 |
| NPAS2 | NANS | 0.761698 | 3 | 0 | 3 |
| NPAS2 | ZNF823 | 0.757917 | 5 | 0 | 5 |
| NPAS2 | ZNF92 | 0.7576 | 3 | 0 | 3 |
| NPAS2 | FRMD6 | 0.743949 | 3 | 0 | 3 |
| NPAS2 | BLNK | 0.737054 | 8 | 0 | 8 |
| NPAS2 | RASGEF1B | 0.731343 | 3 | 0 | 3 |
| NPAS2 | SLC10A7 | 0.729827 | 3 | 0 | 3 |
| NPAS2 | KCTD11 | 0.720755 | 3 | 0 | 3 |
| NPAS2 | MYO6 | 0.718965 | 9 | 0 | 8 |
| NPAS2 | TRIM6 | 0.717751 | 4 | 0 | 4 |
| NPAS2 | TTC9 | 0.715478 | 9 | 0 | 9 |
| NPAS2 | ZNF223 | 0.714481 | 3 | 0 | 3 |
| NPAS2 | DCAF12 | 0.710024 | 3 | 0 | 3 |
| NPAS2 | STX3 | 0.709967 | 4 | 0 | 3 |
| NPAS2 | USP6NL | 0.708535 | 9 | 0 | 7 |
| NPAS2 | UBQLN1 | 0.707464 | 3 | 0 | 3 |
| NPAS2 | EXOC1 | 0.70566 | 8 | 0 | 8 |
For details and further investigation, click here