| Full name: meiosis 1 associated protein | Alias Symbol: D6Mm5e|SPATA37 | ||
| Type: protein-coding gene | Cytoband: 2p13.1 | ||
| Entrez ID: 130951 | HGNC ID: HGNC:25183 | Ensembl Gene: ENSG00000159374 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of M1AP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | M1AP | 130951 | 234498_at | 0.0834 | 0.7852 | |
| GSE26886 | M1AP | 130951 | 234498_at | 0.0861 | 0.3801 | |
| GSE45670 | M1AP | 130951 | 234498_at | 0.0028 | 0.9821 | |
| GSE53622 | M1AP | 130951 | 103351 | 0.9414 | 0.0021 | |
| GSE53624 | M1AP | 130951 | 103351 | 1.0682 | 0.0000 | |
| GSE63941 | M1AP | 130951 | 234498_at | 0.3776 | 0.0117 | |
| GSE77861 | M1AP | 130951 | 234498_at | -0.0796 | 0.5519 | |
| SRP133303 | M1AP | 130951 | RNAseq | 0.9585 | 0.0619 | |
| SRP159526 | M1AP | 130951 | RNAseq | 3.7052 | 0.0000 | |
| SRP193095 | M1AP | 130951 | RNAseq | 1.3716 | 0.0000 |
Upregulated datasets: 3; Downregulated datasets: 0.
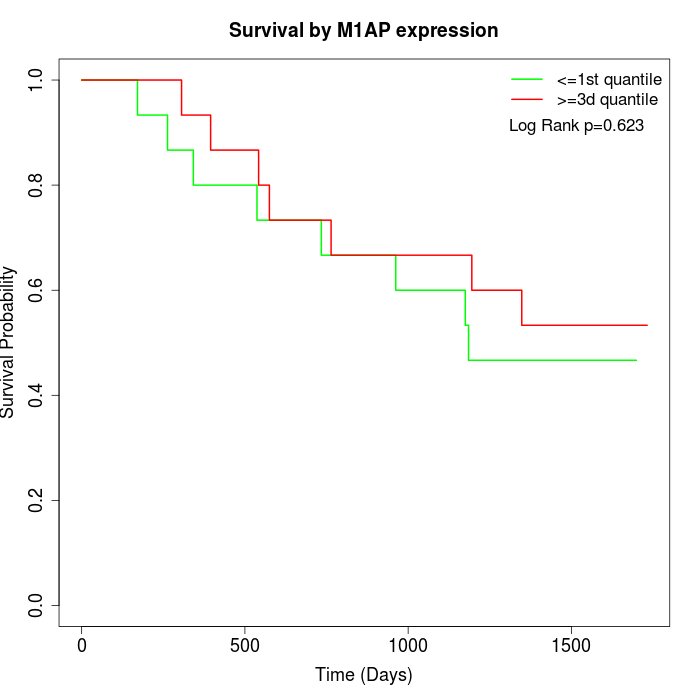
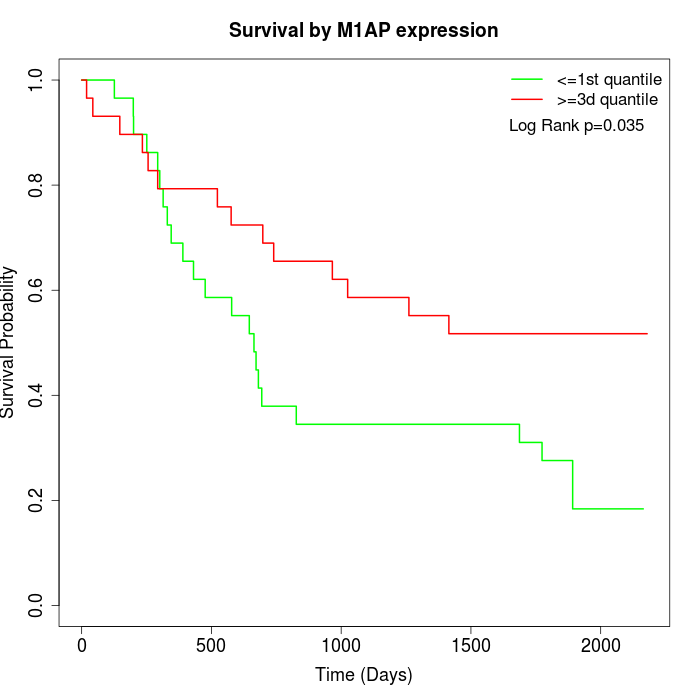
Survival by M1AP expression:
Note: Click image to view full size file.
Copy number change of M1AP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | M1AP | 130951 | 9 | 2 | 19 | |
| GSE20123 | M1AP | 130951 | 10 | 2 | 18 | |
| GSE43470 | M1AP | 130951 | 4 | 0 | 39 | |
| GSE46452 | M1AP | 130951 | 2 | 3 | 54 | |
| GSE47630 | M1AP | 130951 | 7 | 0 | 33 | |
| GSE54993 | M1AP | 130951 | 0 | 6 | 64 | |
| GSE54994 | M1AP | 130951 | 10 | 0 | 43 | |
| GSE60625 | M1AP | 130951 | 0 | 3 | 8 | |
| GSE74703 | M1AP | 130951 | 4 | 0 | 32 | |
| GSE74704 | M1AP | 130951 | 8 | 0 | 12 | |
| TCGA | M1AP | 130951 | 37 | 2 | 57 |
Total number of gains: 91; Total number of losses: 18; Total Number of normals: 379.
Somatic mutations of M1AP:
Generating mutation plots.
Highly correlated genes for M1AP:
Showing top 20/30 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| M1AP | MYCBPAP | 0.664435 | 3 | 0 | 3 |
| M1AP | CELF3 | 0.632067 | 3 | 0 | 3 |
| M1AP | ACSBG2 | 0.620561 | 3 | 0 | 3 |
| M1AP | OGFR | 0.615334 | 3 | 0 | 3 |
| M1AP | BAIAP3 | 0.599462 | 3 | 0 | 3 |
| M1AP | KIR3DL3 | 0.599245 | 3 | 0 | 3 |
| M1AP | ENKUR | 0.587948 | 3 | 0 | 3 |
| M1AP | TTC9B | 0.58757 | 3 | 0 | 3 |
| M1AP | C20orf141 | 0.583409 | 4 | 0 | 4 |
| M1AP | CABP4 | 0.582736 | 3 | 0 | 3 |
| M1AP | NFKBID | 0.581139 | 3 | 0 | 3 |
| M1AP | KCNC3 | 0.572825 | 4 | 0 | 3 |
| M1AP | LINC00558 | 0.571307 | 3 | 0 | 3 |
| M1AP | NFE4 | 0.570212 | 3 | 0 | 3 |
| M1AP | ABCC12 | 0.568613 | 3 | 0 | 3 |
| M1AP | GGN | 0.560127 | 3 | 0 | 3 |
| M1AP | CELA1 | 0.557725 | 4 | 0 | 3 |
| M1AP | MYOG | 0.554061 | 4 | 0 | 3 |
| M1AP | PSG9 | 0.54876 | 4 | 0 | 3 |
| M1AP | GSK3A | 0.54662 | 4 | 0 | 3 |
For details and further investigation, click here