| Full name: gametogenetin | Alias Symbol: FLJ35713|MGC33369 | ||
| Type: protein-coding gene | Cytoband: 19q13.2 | ||
| Entrez ID: 199720 | HGNC ID: HGNC:18869 | Ensembl Gene: ENSG00000179168 | OMIM ID: 609966 |
| Drug and gene relationship at DGIdb | |||
Expression of GGN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GGN | 199720 | 244305_at | 0.0495 | 0.8823 | |
| GSE26886 | GGN | 199720 | 244305_at | 0.2233 | 0.0924 | |
| GSE45670 | GGN | 199720 | 231420_at | 0.1089 | 0.3695 | |
| GSE53622 | GGN | 199720 | 32155 | 0.1511 | 0.0749 | |
| GSE53624 | GGN | 199720 | 32155 | 0.1832 | 0.0063 | |
| GSE63941 | GGN | 199720 | 244305_at | 0.3761 | 0.0090 | |
| GSE77861 | GGN | 199720 | 244305_at | 0.0634 | 0.6912 | |
| GSE97050 | GGN | 199720 | A_33_P3369790 | 0.2026 | 0.3346 | |
| SRP133303 | GGN | 199720 | RNAseq | 0.2405 | 0.3343 | |
| SRP219564 | GGN | 199720 | RNAseq | -0.0476 | 0.8851 | |
| TCGA | GGN | 199720 | RNAseq | 0.3679 | 0.0359 |
Upregulated datasets: 0; Downregulated datasets: 0.
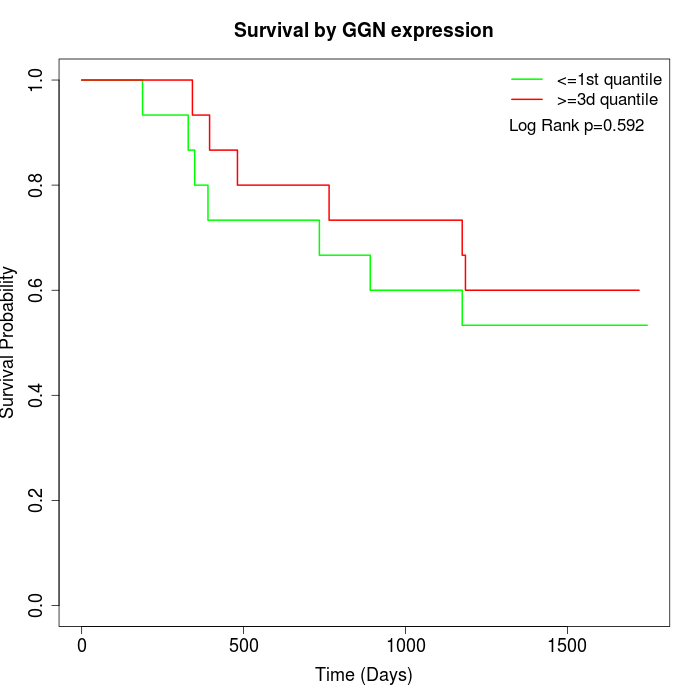
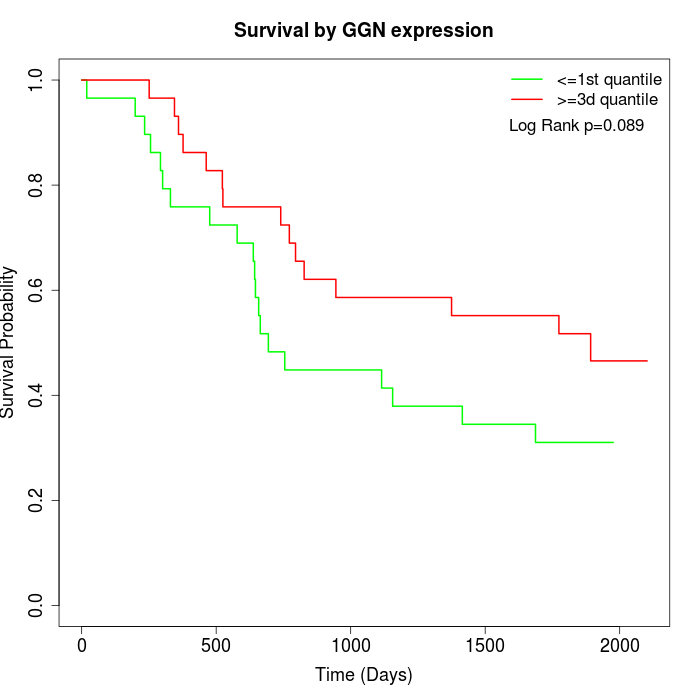
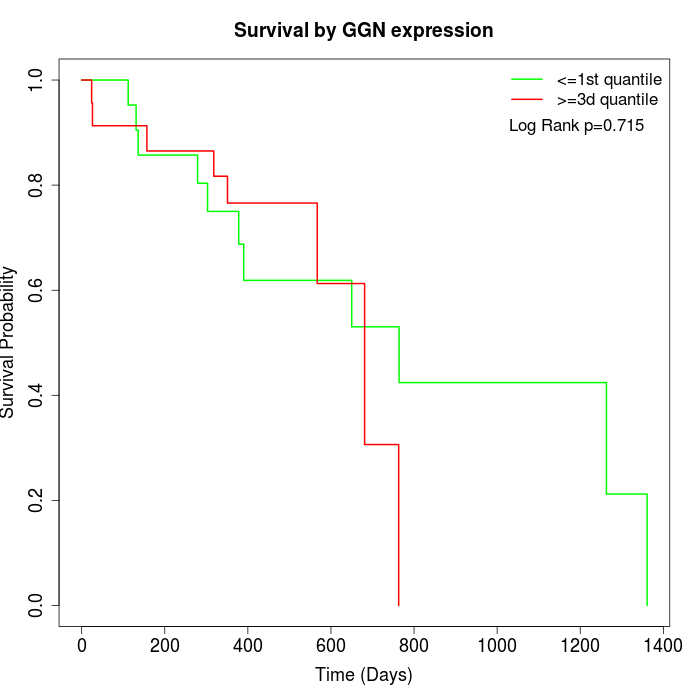
Survival by GGN expression:
Note: Click image to view full size file.
Copy number change of GGN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GGN | 199720 | 7 | 4 | 19 | |
| GSE20123 | GGN | 199720 | 7 | 3 | 20 | |
| GSE43470 | GGN | 199720 | 4 | 10 | 29 | |
| GSE46452 | GGN | 199720 | 47 | 2 | 10 | |
| GSE47630 | GGN | 199720 | 9 | 5 | 26 | |
| GSE54993 | GGN | 199720 | 17 | 3 | 50 | |
| GSE54994 | GGN | 199720 | 6 | 9 | 38 | |
| GSE60625 | GGN | 199720 | 9 | 0 | 2 | |
| GSE74703 | GGN | 199720 | 4 | 6 | 26 | |
| GSE74704 | GGN | 199720 | 7 | 1 | 12 | |
| TCGA | GGN | 199720 | 16 | 15 | 65 |
Total number of gains: 133; Total number of losses: 58; Total Number of normals: 297.
Somatic mutations of GGN:
Generating mutation plots.
Highly correlated genes for GGN:
Showing top 20/323 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GGN | ARHGEF16 | 0.782828 | 3 | 0 | 3 |
| GGN | GTPBP10 | 0.78187 | 3 | 0 | 3 |
| GGN | TMCO6 | 0.774329 | 3 | 0 | 3 |
| GGN | CTF1 | 0.763206 | 3 | 0 | 3 |
| GGN | PA2G4 | 0.760466 | 3 | 0 | 3 |
| GGN | KCNC4 | 0.757825 | 4 | 0 | 4 |
| GGN | ABHD1 | 0.756761 | 3 | 0 | 3 |
| GGN | DHPS | 0.755187 | 3 | 0 | 3 |
| GGN | TRAPPC1 | 0.749567 | 3 | 0 | 3 |
| GGN | DDIT3 | 0.748063 | 3 | 0 | 3 |
| GGN | FBXL6 | 0.744907 | 4 | 0 | 4 |
| GGN | PRKCSH | 0.741221 | 3 | 0 | 3 |
| GGN | PRRC2A | 0.738812 | 4 | 0 | 4 |
| GGN | C12orf45 | 0.738331 | 3 | 0 | 3 |
| GGN | SPTAN1 | 0.737504 | 3 | 0 | 3 |
| GGN | ZNF408 | 0.734749 | 3 | 0 | 3 |
| GGN | FNDC8 | 0.734717 | 3 | 0 | 3 |
| GGN | RHAG | 0.732169 | 3 | 0 | 3 |
| GGN | FAM86C1 | 0.730556 | 3 | 0 | 3 |
| GGN | ACIN1 | 0.730446 | 3 | 0 | 3 |
For details and further investigation, click here