| Full name: nebulin related anchoring protein | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q25.3 | ||
| Entrez ID: 4892 | HGNC ID: HGNC:7988 | Ensembl Gene: ENSG00000197893 | OMIM ID: 602873 |
| Drug and gene relationship at DGIdb | |||
Expression of NRAP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NRAP | 4892 | 235312_s_at | -0.0350 | 0.9218 | |
| GSE20347 | NRAP | 4892 | 207089_at | -0.0425 | 0.5306 | |
| GSE23400 | NRAP | 4892 | 207089_at | -0.0147 | 0.6236 | |
| GSE26886 | NRAP | 4892 | 235312_s_at | -0.2579 | 0.0825 | |
| GSE29001 | NRAP | 4892 | 207089_at | 0.0011 | 0.9915 | |
| GSE38129 | NRAP | 4892 | 207089_at | -0.0709 | 0.2229 | |
| GSE45670 | NRAP | 4892 | 235312_s_at | 0.0440 | 0.6356 | |
| GSE63941 | NRAP | 4892 | 235312_s_at | 0.2326 | 0.1284 | |
| GSE77861 | NRAP | 4892 | 235313_at | -0.3933 | 0.0096 | |
| GSE97050 | NRAP | 4892 | A_23_P402765 | -1.4057 | 0.2162 | |
| TCGA | NRAP | 4892 | RNAseq | -3.7709 | 0.0071 |
Upregulated datasets: 0; Downregulated datasets: 1.
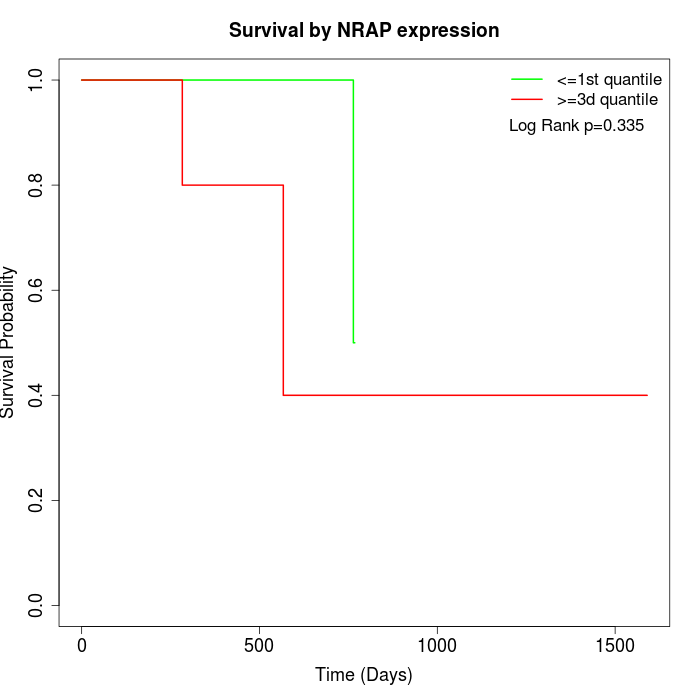
Survival by NRAP expression:
Note: Click image to view full size file.
Copy number change of NRAP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NRAP | 4892 | 1 | 8 | 21 | |
| GSE20123 | NRAP | 4892 | 1 | 7 | 22 | |
| GSE43470 | NRAP | 4892 | 0 | 9 | 34 | |
| GSE46452 | NRAP | 4892 | 0 | 11 | 48 | |
| GSE47630 | NRAP | 4892 | 2 | 14 | 24 | |
| GSE54993 | NRAP | 4892 | 8 | 0 | 62 | |
| GSE54994 | NRAP | 4892 | 1 | 10 | 42 | |
| GSE60625 | NRAP | 4892 | 0 | 0 | 11 | |
| GSE74703 | NRAP | 4892 | 0 | 6 | 30 | |
| GSE74704 | NRAP | 4892 | 1 | 3 | 16 | |
| TCGA | NRAP | 4892 | 4 | 28 | 64 |
Total number of gains: 18; Total number of losses: 96; Total Number of normals: 374.
Somatic mutations of NRAP:
Generating mutation plots.
Highly correlated genes for NRAP:
Showing top 20/386 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NRAP | LEMD1-AS1 | 0.714514 | 3 | 0 | 3 |
| NRAP | ANHX | 0.678928 | 4 | 0 | 3 |
| NRAP | PNPLA4 | 0.677105 | 3 | 0 | 3 |
| NRAP | MRGPRX2 | 0.67333 | 5 | 0 | 4 |
| NRAP | MAPK12 | 0.65871 | 3 | 0 | 3 |
| NRAP | GABRG1 | 0.649713 | 3 | 0 | 3 |
| NRAP | OR51M1 | 0.649286 | 5 | 0 | 4 |
| NRAP | LINC00330 | 0.64393 | 3 | 0 | 3 |
| NRAP | STPG2 | 0.640599 | 4 | 0 | 3 |
| NRAP | ADIG | 0.639483 | 4 | 0 | 4 |
| NRAP | AMER2 | 0.637894 | 3 | 0 | 3 |
| NRAP | ARX | 0.637399 | 4 | 0 | 3 |
| NRAP | HCG22 | 0.633454 | 3 | 0 | 3 |
| NRAP | CPS1-IT1 | 0.63316 | 3 | 0 | 3 |
| NRAP | LINC01192 | 0.632715 | 5 | 0 | 4 |
| NRAP | LINC01210 | 0.631768 | 3 | 0 | 3 |
| NRAP | LINC00930 | 0.629446 | 4 | 0 | 4 |
| NRAP | NFIA-AS2 | 0.628909 | 3 | 0 | 3 |
| NRAP | ANKRD31 | 0.623598 | 4 | 0 | 3 |
| NRAP | PF4 | 0.61871 | 4 | 0 | 4 |
For details and further investigation, click here