| Full name: gamma-aminobutyric acid type A receptor gamma1 subunit | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 4p12 | ||
| Entrez ID: 2565 | HGNC ID: HGNC:4086 | Ensembl Gene: ENSG00000163285 | OMIM ID: 137166 |
| Drug and gene relationship at DGIdb | |||
GABRG1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04727 | GABAergic synapse | |
| hsa05032 | Morphine addiction |
Expression of GABRG1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GABRG1 | 2565 | 1552943_at | -0.0413 | 0.8743 | |
| GSE26886 | GABRG1 | 2565 | 241805_at | 0.0061 | 0.9608 | |
| GSE45670 | GABRG1 | 2565 | 241805_at | 0.0374 | 0.6620 | |
| GSE53622 | GABRG1 | 2565 | 12823 | -0.4068 | 0.0050 | |
| GSE53624 | GABRG1 | 2565 | 12823 | -0.4886 | 0.0000 | |
| GSE63941 | GABRG1 | 2565 | 241805_at | 0.1225 | 0.4996 | |
| GSE77861 | GABRG1 | 2565 | 241805_at | -0.0951 | 0.2503 | |
| TCGA | GABRG1 | 2565 | RNAseq | -1.9747 | 0.1569 |
Upregulated datasets: 0; Downregulated datasets: 0.
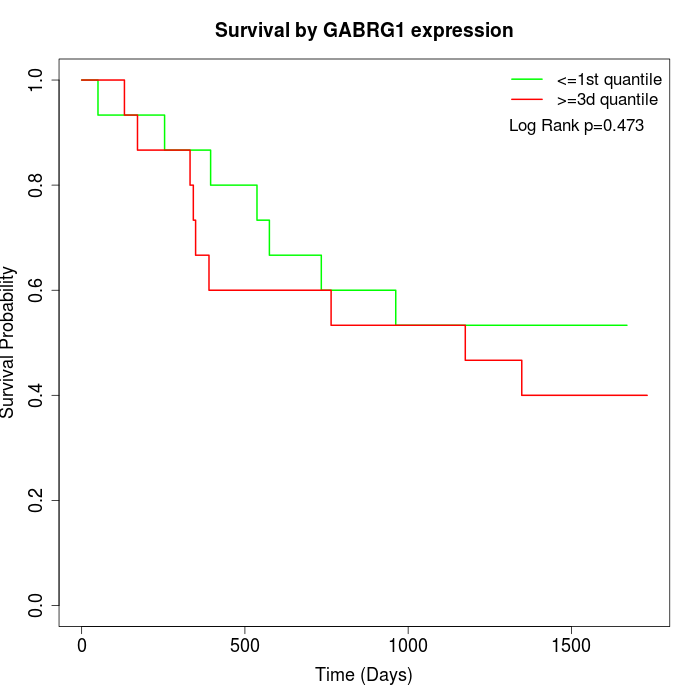
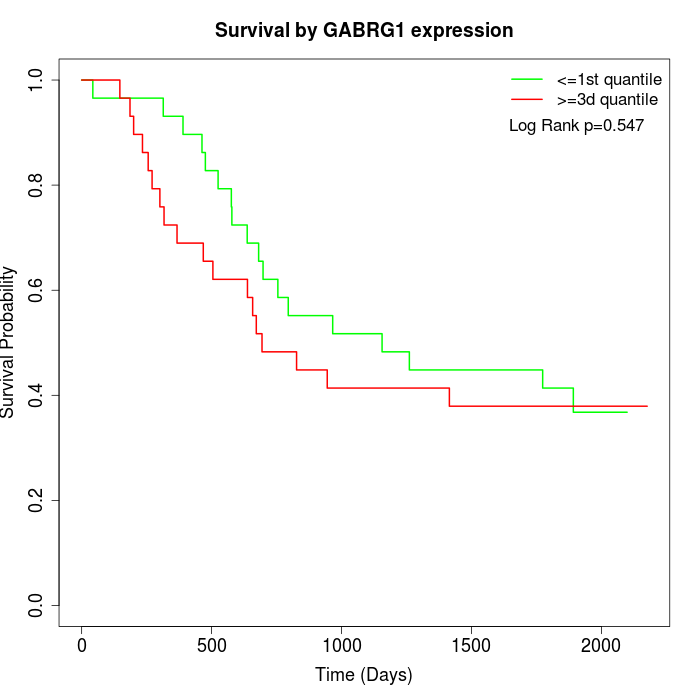
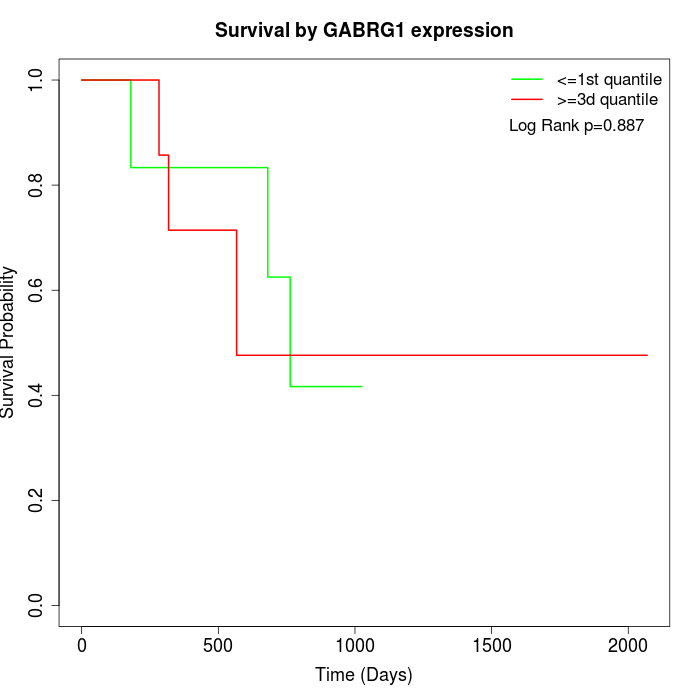
Survival by GABRG1 expression:
Note: Click image to view full size file.
Copy number change of GABRG1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GABRG1 | 2565 | 0 | 16 | 14 | |
| GSE20123 | GABRG1 | 2565 | 0 | 16 | 14 | |
| GSE43470 | GABRG1 | 2565 | 0 | 14 | 29 | |
| GSE46452 | GABRG1 | 2565 | 1 | 37 | 21 | |
| GSE47630 | GABRG1 | 2565 | 1 | 18 | 21 | |
| GSE54993 | GABRG1 | 2565 | 7 | 0 | 63 | |
| GSE54994 | GABRG1 | 2565 | 4 | 9 | 40 | |
| GSE60625 | GABRG1 | 2565 | 0 | 0 | 11 | |
| GSE74703 | GABRG1 | 2565 | 0 | 12 | 24 | |
| GSE74704 | GABRG1 | 2565 | 0 | 9 | 11 | |
| TCGA | GABRG1 | 2565 | 15 | 35 | 46 |
Total number of gains: 28; Total number of losses: 166; Total Number of normals: 294.
Somatic mutations of GABRG1:
Generating mutation plots.
Highly correlated genes for GABRG1:
Showing top 20/24 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GABRG1 | ZNF214 | 0.67243 | 3 | 0 | 3 |
| GABRG1 | NRAP | 0.649713 | 3 | 0 | 3 |
| GABRG1 | BSND | 0.620157 | 3 | 0 | 3 |
| GABRG1 | DPEP3 | 0.615377 | 3 | 0 | 3 |
| GABRG1 | LINC00937 | 0.591173 | 3 | 0 | 3 |
| GABRG1 | OLIG3 | 0.580169 | 3 | 0 | 3 |
| GABRG1 | SUN5 | 0.576824 | 3 | 0 | 3 |
| GABRG1 | OR51M1 | 0.570263 | 4 | 0 | 3 |
| GABRG1 | SCARNA2 | 0.569402 | 3 | 0 | 3 |
| GABRG1 | CLVS1 | 0.569157 | 4 | 0 | 3 |
| GABRG1 | RBPJL | 0.561503 | 4 | 0 | 3 |
| GABRG1 | XAGE2 | 0.561239 | 4 | 0 | 3 |
| GABRG1 | CELA3B | 0.55994 | 3 | 0 | 3 |
| GABRG1 | KLK4 | 0.556357 | 5 | 0 | 4 |
| GABRG1 | SAMD10 | 0.555767 | 3 | 0 | 3 |
| GABRG1 | FAM9B | 0.53707 | 4 | 0 | 3 |
| GABRG1 | RTP3 | 0.534235 | 3 | 0 | 3 |
| GABRG1 | TBC1D21 | 0.531564 | 3 | 0 | 3 |
| GABRG1 | RPPH1 | 0.529814 | 3 | 0 | 3 |
| GABRG1 | PLIN5 | 0.526482 | 4 | 0 | 3 |
For details and further investigation, click here