| Full name: SREBF chaperone | Alias Symbol: KIAA0199 | ||
| Type: protein-coding gene | Cytoband: 3p21.31 | ||
| Entrez ID: 22937 | HGNC ID: HGNC:30634 | Ensembl Gene: ENSG00000114650 | OMIM ID: 601510 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of SCAP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SCAP | 22937 | 212329_at | 0.2004 | 0.5947 | |
| GSE20347 | SCAP | 22937 | 212329_at | -0.5135 | 0.0005 | |
| GSE23400 | SCAP | 22937 | 212329_at | -0.2225 | 0.0002 | |
| GSE26886 | SCAP | 22937 | 212329_at | -0.4765 | 0.0167 | |
| GSE29001 | SCAP | 22937 | 212329_at | -0.3709 | 0.0338 | |
| GSE38129 | SCAP | 22937 | 212329_at | -0.4104 | 0.0000 | |
| GSE45670 | SCAP | 22937 | 212329_at | -0.1381 | 0.2344 | |
| GSE53622 | SCAP | 22937 | 21361 | -0.4058 | 0.0000 | |
| GSE53624 | SCAP | 22937 | 21361 | -0.3984 | 0.0000 | |
| GSE63941 | SCAP | 22937 | 212329_at | -0.9394 | 0.0040 | |
| GSE77861 | SCAP | 22937 | 212329_at | -0.2266 | 0.3199 | |
| GSE97050 | SCAP | 22937 | A_23_P252825 | -0.1997 | 0.3406 | |
| SRP007169 | SCAP | 22937 | RNAseq | -0.9786 | 0.0135 | |
| SRP008496 | SCAP | 22937 | RNAseq | -1.0018 | 0.0000 | |
| SRP064894 | SCAP | 22937 | RNAseq | -0.1270 | 0.4667 | |
| SRP133303 | SCAP | 22937 | RNAseq | -0.7385 | 0.0000 | |
| SRP159526 | SCAP | 22937 | RNAseq | -0.4472 | 0.0800 | |
| SRP193095 | SCAP | 22937 | RNAseq | -0.4197 | 0.0000 | |
| SRP219564 | SCAP | 22937 | RNAseq | -0.1901 | 0.6774 | |
| TCGA | SCAP | 22937 | RNAseq | -0.1544 | 0.0007 |
Upregulated datasets: 0; Downregulated datasets: 1.
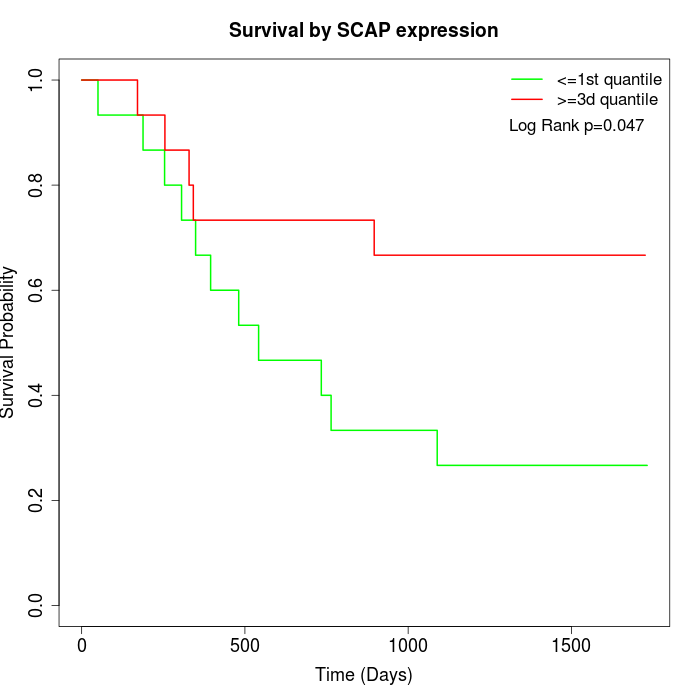
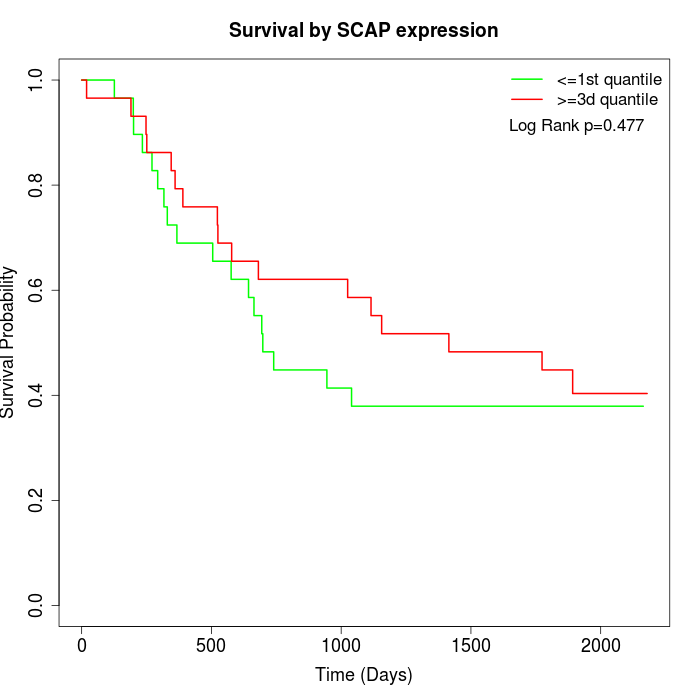
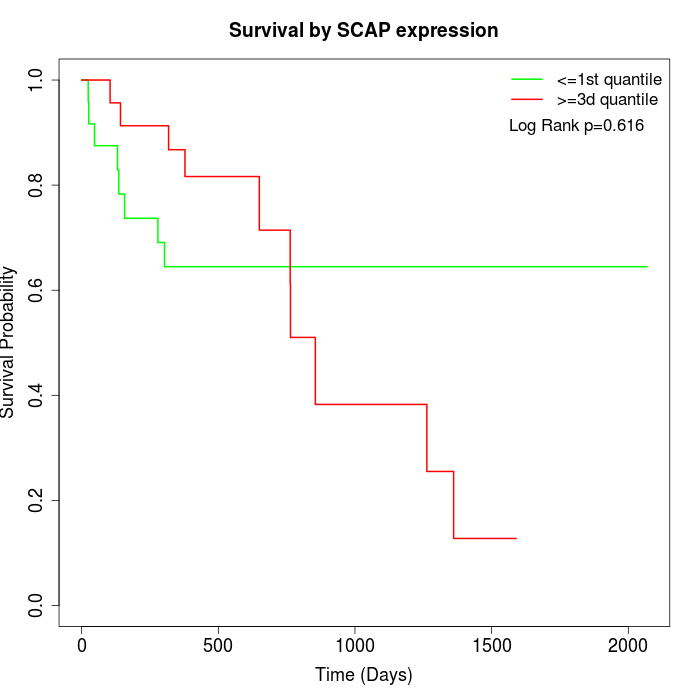
Survival by SCAP expression:
Note: Click image to view full size file.
Copy number change of SCAP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SCAP | 22937 | 0 | 18 | 12 | |
| GSE20123 | SCAP | 22937 | 0 | 19 | 11 | |
| GSE43470 | SCAP | 22937 | 0 | 19 | 24 | |
| GSE46452 | SCAP | 22937 | 2 | 17 | 40 | |
| GSE47630 | SCAP | 22937 | 2 | 23 | 15 | |
| GSE54993 | SCAP | 22937 | 7 | 2 | 61 | |
| GSE54994 | SCAP | 22937 | 0 | 36 | 17 | |
| GSE60625 | SCAP | 22937 | 5 | 0 | 6 | |
| GSE74703 | SCAP | 22937 | 0 | 15 | 21 | |
| GSE74704 | SCAP | 22937 | 0 | 12 | 8 | |
| TCGA | SCAP | 22937 | 1 | 75 | 20 |
Total number of gains: 17; Total number of losses: 236; Total Number of normals: 235.
Somatic mutations of SCAP:
Generating mutation plots.
Highly correlated genes for SCAP:
Showing top 20/179 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SCAP | SHROOM4 | 0.641089 | 3 | 0 | 3 |
| SCAP | MAPKAPK3 | 0.634481 | 11 | 0 | 10 |
| SCAP | MTMR10 | 0.630501 | 4 | 0 | 4 |
| SCAP | HYMAI | 0.623838 | 4 | 0 | 3 |
| SCAP | DNM2 | 0.622567 | 3 | 0 | 3 |
| SCAP | RAB11B-AS1 | 0.618543 | 3 | 0 | 3 |
| SCAP | WDR82 | 0.616852 | 8 | 0 | 6 |
| SCAP | RPL14 | 0.61286 | 7 | 0 | 6 |
| SCAP | DHX34 | 0.609596 | 3 | 0 | 3 |
| SCAP | KRTAP20-1 | 0.605089 | 3 | 0 | 3 |
| SCAP | BTBD19 | 0.604096 | 3 | 0 | 3 |
| SCAP | VARS2 | 0.602604 | 7 | 0 | 5 |
| SCAP | COX19 | 0.599972 | 3 | 0 | 3 |
| SCAP | SFI1 | 0.592679 | 4 | 0 | 3 |
| SCAP | FAM163B | 0.592205 | 3 | 0 | 3 |
| SCAP | SRCAP | 0.587342 | 5 | 0 | 3 |
| SCAP | SIL1 | 0.582552 | 3 | 0 | 3 |
| SCAP | MAFG | 0.582311 | 4 | 0 | 3 |
| SCAP | CC2D1B | 0.58136 | 5 | 0 | 3 |
| SCAP | SH3BGRL3 | 0.578532 | 6 | 0 | 5 |
For details and further investigation, click here