| Full name: dynamin 2 | Alias Symbol: DYNII|DYN2|CMTDIB|CMTDI1|DI-CMTB|CMT2M | ||
| Type: protein-coding gene | Cytoband: 19p13.2 | ||
| Entrez ID: 1785 | HGNC ID: HGNC:2974 | Ensembl Gene: ENSG00000079805 | OMIM ID: 602378 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
DNM2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04666 | Fc gamma R-mediated phagocytosis | |
| hsa05100 | Bacterial invasion of epithelial cells |
Expression of DNM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DNM2 | 1785 | 202253_s_at | 0.2048 | 0.7402 | |
| GSE20347 | DNM2 | 1785 | 202253_s_at | -0.2368 | 0.1053 | |
| GSE23400 | DNM2 | 1785 | 202253_s_at | -0.0643 | 0.3112 | |
| GSE26886 | DNM2 | 1785 | 202253_s_at | -0.0297 | 0.8877 | |
| GSE29001 | DNM2 | 1785 | 202253_s_at | -0.2799 | 0.5542 | |
| GSE38129 | DNM2 | 1785 | 202253_s_at | -0.0992 | 0.4386 | |
| GSE45670 | DNM2 | 1785 | 202253_s_at | 0.1448 | 0.2071 | |
| GSE53622 | DNM2 | 1785 | 50661 | -0.0901 | 0.0629 | |
| GSE53624 | DNM2 | 1785 | 125924 | -0.2646 | 0.0000 | |
| GSE63941 | DNM2 | 1785 | 202253_s_at | 0.8696 | 0.0072 | |
| GSE77861 | DNM2 | 1785 | 202253_s_at | -0.0102 | 0.9643 | |
| GSE97050 | DNM2 | 1785 | A_23_P407074 | -0.5640 | 0.5305 | |
| SRP007169 | DNM2 | 1785 | RNAseq | -0.7235 | 0.0987 | |
| SRP008496 | DNM2 | 1785 | RNAseq | -0.7789 | 0.0021 | |
| SRP064894 | DNM2 | 1785 | RNAseq | -0.2529 | 0.0863 | |
| SRP133303 | DNM2 | 1785 | RNAseq | -0.2643 | 0.0532 | |
| SRP159526 | DNM2 | 1785 | RNAseq | -0.5603 | 0.0001 | |
| SRP193095 | DNM2 | 1785 | RNAseq | -0.4988 | 0.0032 | |
| SRP219564 | DNM2 | 1785 | RNAseq | -0.0825 | 0.7616 | |
| TCGA | DNM2 | 1785 | RNAseq | -0.1388 | 0.0004 |
Upregulated datasets: 0; Downregulated datasets: 0.
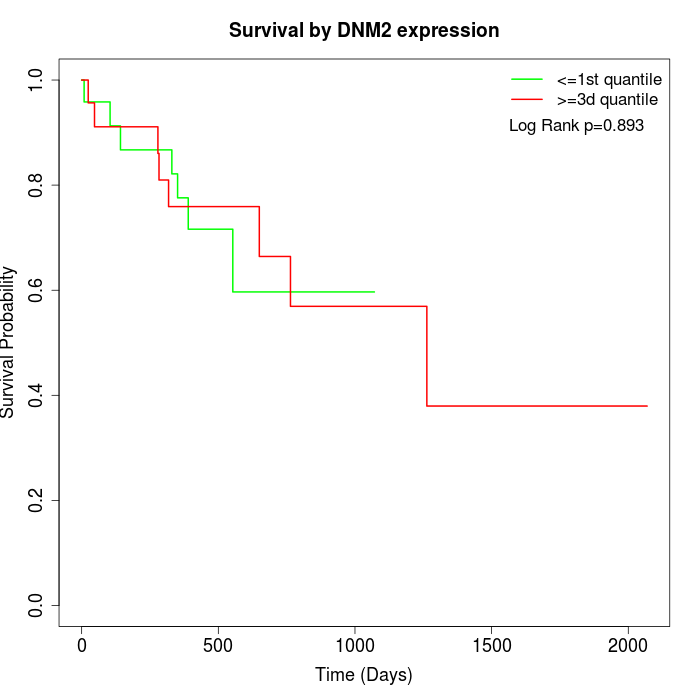
Survival by DNM2 expression:
Note: Click image to view full size file.
Copy number change of DNM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DNM2 | 1785 | 4 | 3 | 23 | |
| GSE20123 | DNM2 | 1785 | 3 | 1 | 26 | |
| GSE43470 | DNM2 | 1785 | 3 | 6 | 34 | |
| GSE46452 | DNM2 | 1785 | 47 | 1 | 11 | |
| GSE47630 | DNM2 | 1785 | 4 | 7 | 29 | |
| GSE54993 | DNM2 | 1785 | 16 | 3 | 51 | |
| GSE54994 | DNM2 | 1785 | 6 | 13 | 34 | |
| GSE60625 | DNM2 | 1785 | 9 | 0 | 2 | |
| GSE74703 | DNM2 | 1785 | 3 | 4 | 29 | |
| GSE74704 | DNM2 | 1785 | 0 | 1 | 19 | |
| TCGA | DNM2 | 1785 | 15 | 14 | 67 |
Total number of gains: 110; Total number of losses: 53; Total Number of normals: 325.
Somatic mutations of DNM2:
Generating mutation plots.
Highly correlated genes for DNM2:
Showing top 20/196 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DNM2 | LRCH4 | 0.701898 | 3 | 0 | 3 |
| DNM2 | PCIF1 | 0.674126 | 3 | 0 | 3 |
| DNM2 | SPTAN1 | 0.666269 | 4 | 0 | 4 |
| DNM2 | KCTD17 | 0.665634 | 3 | 0 | 3 |
| DNM2 | SNAPC2 | 0.655 | 3 | 0 | 3 |
| DNM2 | PODXL2 | 0.654439 | 4 | 0 | 4 |
| DNM2 | SURF2 | 0.652295 | 3 | 0 | 3 |
| DNM2 | NFYC | 0.649875 | 4 | 0 | 3 |
| DNM2 | TSEN54 | 0.636617 | 3 | 0 | 3 |
| DNM2 | PFN1 | 0.636162 | 3 | 0 | 3 |
| DNM2 | CC2D1A | 0.631909 | 9 | 0 | 8 |
| DNM2 | IGFBPL1 | 0.631168 | 3 | 0 | 3 |
| DNM2 | COPS7A | 0.628156 | 3 | 0 | 3 |
| DNM2 | SCAP | 0.622567 | 3 | 0 | 3 |
| DNM2 | RFNG | 0.620755 | 4 | 0 | 3 |
| DNM2 | NDUFA11 | 0.619697 | 3 | 0 | 3 |
| DNM2 | FBXL19 | 0.611643 | 4 | 0 | 3 |
| DNM2 | QTRT1 | 0.609825 | 5 | 0 | 4 |
| DNM2 | MIIP | 0.60861 | 6 | 0 | 5 |
| DNM2 | CHMP6 | 0.605147 | 3 | 0 | 3 |
For details and further investigation, click here