| Full name: ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12q14.1 | ||
| Entrez ID: 116986 | HGNC ID: HGNC:16921 | Ensembl Gene: ENSG00000135439 | OMIM ID: 605476 |
| Drug and gene relationship at DGIdb | |||
AGAP2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04068 | FoxO signaling pathway |
Expression of AGAP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | AGAP2 | 116986 | 206152_at | 0.0854 | 0.7240 | |
| GSE20347 | AGAP2 | 116986 | 206152_at | 0.0262 | 0.7674 | |
| GSE23400 | AGAP2 | 116986 | 206152_at | -0.0662 | 0.0319 | |
| GSE26886 | AGAP2 | 116986 | 206152_at | 0.3006 | 0.0037 | |
| GSE29001 | AGAP2 | 116986 | 206152_at | -0.0370 | 0.8695 | |
| GSE38129 | AGAP2 | 116986 | 206152_at | -0.1063 | 0.1047 | |
| GSE45670 | AGAP2 | 116986 | 206152_at | 0.0594 | 0.4904 | |
| GSE53622 | AGAP2 | 116986 | 1270 | 0.0278 | 0.7345 | |
| GSE53624 | AGAP2 | 116986 | 1270 | 0.0307 | 0.6744 | |
| GSE63941 | AGAP2 | 116986 | 206152_at | 0.2887 | 0.0492 | |
| GSE77861 | AGAP2 | 116986 | 206152_at | -0.0744 | 0.6531 | |
| GSE97050 | AGAP2 | 116986 | A_33_P3352148 | 0.1602 | 0.4492 | |
| SRP064894 | AGAP2 | 116986 | RNAseq | 0.6108 | 0.0217 | |
| SRP133303 | AGAP2 | 116986 | RNAseq | -0.1832 | 0.3522 | |
| SRP159526 | AGAP2 | 116986 | RNAseq | -0.6137 | 0.3087 | |
| SRP193095 | AGAP2 | 116986 | RNAseq | 0.3766 | 0.0593 | |
| SRP219564 | AGAP2 | 116986 | RNAseq | -0.0195 | 0.9573 | |
| TCGA | AGAP2 | 116986 | RNAseq | 0.0156 | 0.9207 |
Upregulated datasets: 0; Downregulated datasets: 0.
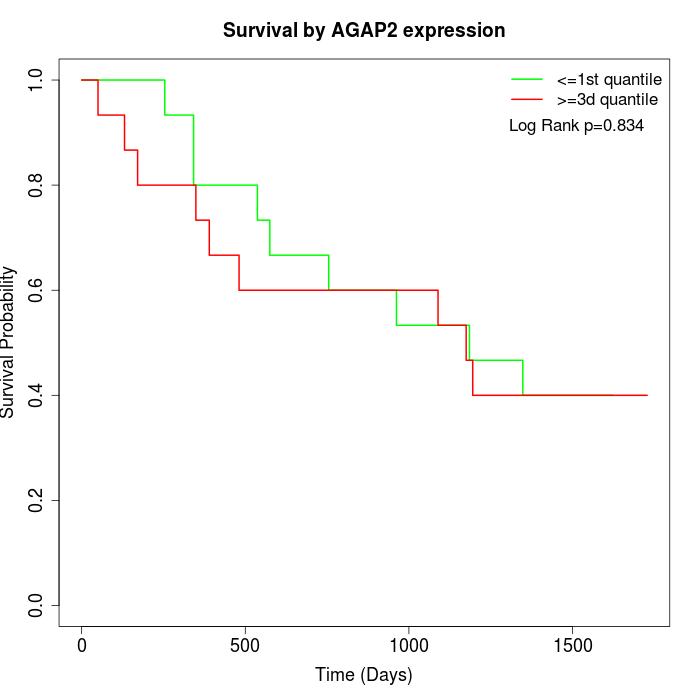
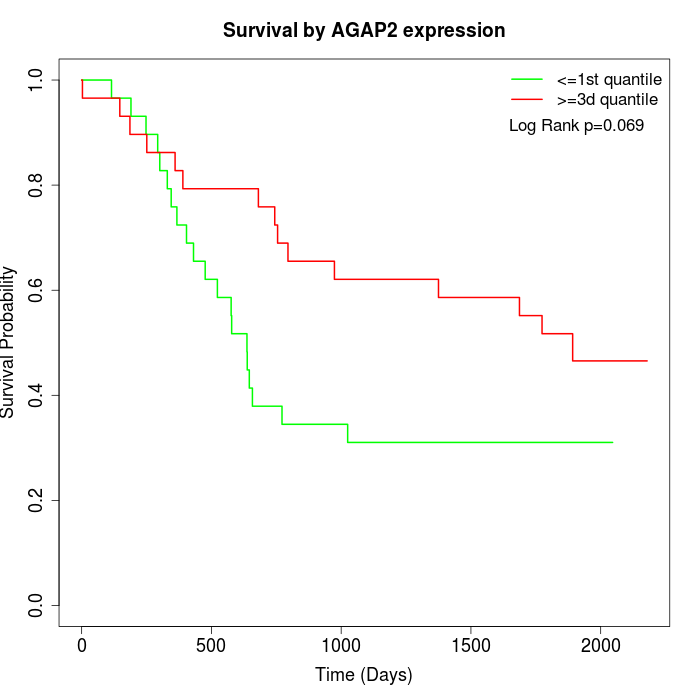
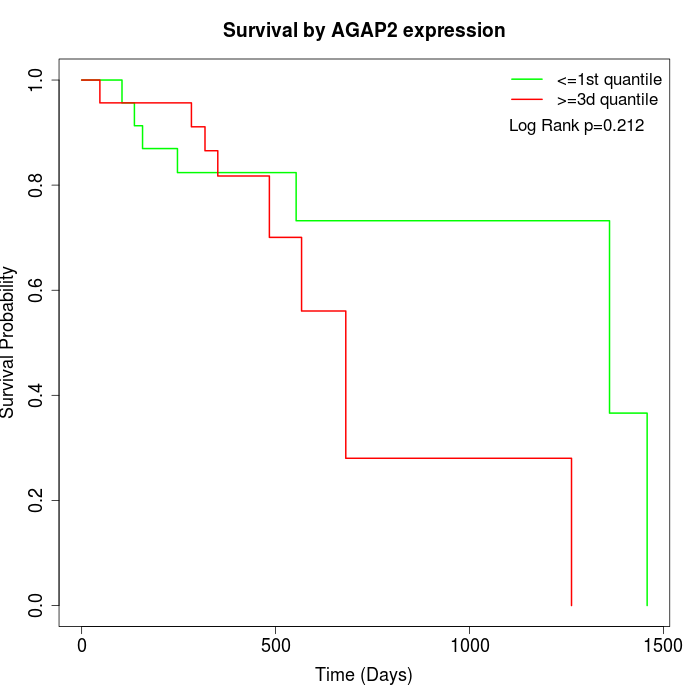
Survival by AGAP2 expression:
Note: Click image to view full size file.
Copy number change of AGAP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AGAP2 | 116986 | 7 | 1 | 22 | |
| GSE20123 | AGAP2 | 116986 | 7 | 0 | 23 | |
| GSE43470 | AGAP2 | 116986 | 2 | 0 | 41 | |
| GSE46452 | AGAP2 | 116986 | 8 | 1 | 50 | |
| GSE47630 | AGAP2 | 116986 | 10 | 2 | 28 | |
| GSE54993 | AGAP2 | 116986 | 0 | 5 | 65 | |
| GSE54994 | AGAP2 | 116986 | 4 | 1 | 48 | |
| GSE60625 | AGAP2 | 116986 | 0 | 0 | 11 | |
| GSE74703 | AGAP2 | 116986 | 2 | 0 | 34 | |
| GSE74704 | AGAP2 | 116986 | 5 | 0 | 15 | |
| TCGA | AGAP2 | 116986 | 14 | 10 | 72 |
Total number of gains: 59; Total number of losses: 20; Total Number of normals: 409.
Somatic mutations of AGAP2:
Generating mutation plots.
Highly correlated genes for AGAP2:
Showing top 20/782 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AGAP2 | FAM178B | 0.818104 | 3 | 0 | 3 |
| AGAP2 | NXNL1 | 0.801898 | 3 | 0 | 3 |
| AGAP2 | TMEM221 | 0.778041 | 3 | 0 | 3 |
| AGAP2 | GAST | 0.770886 | 3 | 0 | 3 |
| AGAP2 | GPC2 | 0.768784 | 3 | 0 | 3 |
| AGAP2 | LENG1 | 0.751664 | 3 | 0 | 3 |
| AGAP2 | BTBD8 | 0.745136 | 3 | 0 | 3 |
| AGAP2 | KLHL35 | 0.744064 | 3 | 0 | 3 |
| AGAP2 | CDK16 | 0.742706 | 3 | 0 | 3 |
| AGAP2 | C1QTNF6 | 0.741976 | 3 | 0 | 3 |
| AGAP2 | C1QL1 | 0.741749 | 4 | 0 | 4 |
| AGAP2 | SPATA2L | 0.736548 | 3 | 0 | 3 |
| AGAP2 | BCL2L10 | 0.733898 | 4 | 0 | 4 |
| AGAP2 | OR10H1 | 0.731809 | 4 | 0 | 3 |
| AGAP2 | IGSF9B | 0.731379 | 4 | 0 | 3 |
| AGAP2 | SCN1B | 0.729739 | 4 | 0 | 3 |
| AGAP2 | RPP25 | 0.725913 | 3 | 0 | 3 |
| AGAP2 | MYOD1 | 0.723795 | 4 | 0 | 3 |
| AGAP2 | PNMA6A | 0.722749 | 4 | 0 | 4 |
| AGAP2 | KIF17 | 0.71718 | 4 | 0 | 4 |
For details and further investigation, click here