| Full name: kelch like family member 35 | Alias Symbol: FLJ33790 | ||
| Type: protein-coding gene | Cytoband: 11q13.4 | ||
| Entrez ID: 283212 | HGNC ID: HGNC:26597 | Ensembl Gene: ENSG00000149243 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of KLHL35:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KLHL35 | 283212 | 1553611_s_at | 0.3717 | 0.4589 | |
| GSE20347 | KLHL35 | 283212 | 215748_at | 0.0131 | 0.9069 | |
| GSE23400 | KLHL35 | 283212 | 214208_at | -0.0828 | 0.0202 | |
| GSE26886 | KLHL35 | 283212 | 1553611_s_at | 0.2580 | 0.4825 | |
| GSE29001 | KLHL35 | 283212 | 215748_at | -0.0402 | 0.8174 | |
| GSE38129 | KLHL35 | 283212 | 215748_at | -0.0972 | 0.2361 | |
| GSE45670 | KLHL35 | 283212 | 1553611_s_at | 0.1818 | 0.1273 | |
| GSE53622 | KLHL35 | 283212 | 18836 | 0.3549 | 0.0009 | |
| GSE53624 | KLHL35 | 283212 | 18836 | 0.6861 | 0.0000 | |
| GSE63941 | KLHL35 | 283212 | 1553611_s_at | -0.1432 | 0.6647 | |
| GSE77861 | KLHL35 | 283212 | 1553611_s_at | -0.0649 | 0.6147 | |
| GSE97050 | KLHL35 | 283212 | A_23_P351837 | 0.1047 | 0.6195 | |
| SRP133303 | KLHL35 | 283212 | RNAseq | 0.0274 | 0.9138 | |
| SRP219564 | KLHL35 | 283212 | RNAseq | 2.6351 | 0.0005 | |
| TCGA | KLHL35 | 283212 | RNAseq | -0.0743 | 0.8310 |
Upregulated datasets: 1; Downregulated datasets: 0.
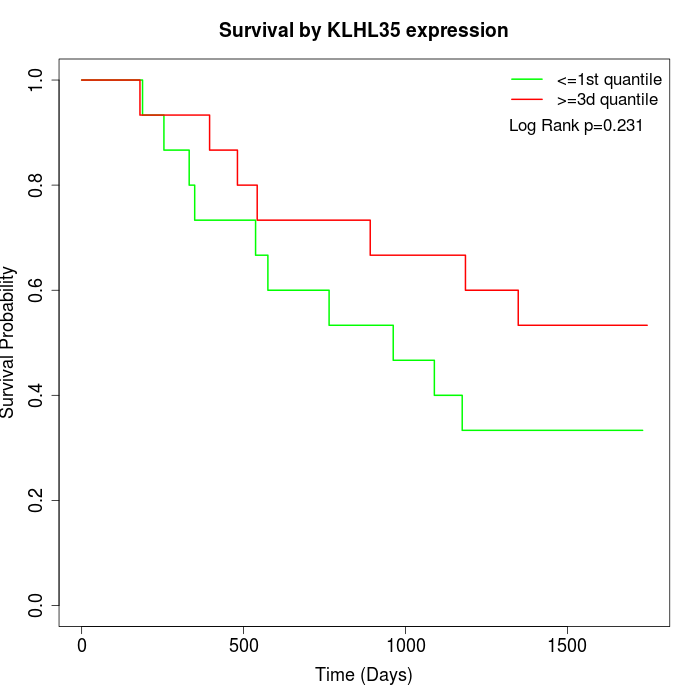
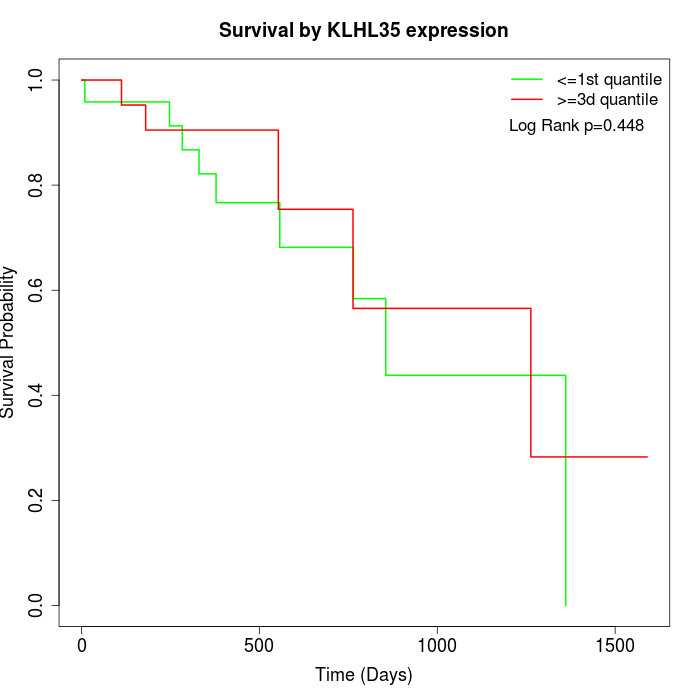
Survival by KLHL35 expression:
Note: Click image to view full size file.
Copy number change of KLHL35:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KLHL35 | 283212 | 6 | 8 | 16 | |
| GSE20123 | KLHL35 | 283212 | 6 | 7 | 17 | |
| GSE43470 | KLHL35 | 283212 | 5 | 4 | 34 | |
| GSE46452 | KLHL35 | 283212 | 7 | 22 | 30 | |
| GSE47630 | KLHL35 | 283212 | 6 | 14 | 20 | |
| GSE54993 | KLHL35 | 283212 | 8 | 2 | 60 | |
| GSE54994 | KLHL35 | 283212 | 7 | 16 | 30 | |
| GSE60625 | KLHL35 | 283212 | 4 | 3 | 4 | |
| GSE74703 | KLHL35 | 283212 | 4 | 3 | 29 | |
| GSE74704 | KLHL35 | 283212 | 4 | 3 | 13 | |
| TCGA | KLHL35 | 283212 | 21 | 32 | 43 |
Total number of gains: 78; Total number of losses: 114; Total Number of normals: 296.
Somatic mutations of KLHL35:
Generating mutation plots.
Highly correlated genes for KLHL35:
Showing top 20/444 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KLHL35 | SCGN | 0.818986 | 3 | 0 | 3 |
| KLHL35 | LIMD1 | 0.787487 | 4 | 0 | 3 |
| KLHL35 | GBX2 | 0.7869 | 3 | 0 | 3 |
| KLHL35 | CPLX2 | 0.780804 | 3 | 0 | 3 |
| KLHL35 | BAZ2A | 0.748666 | 3 | 0 | 3 |
| KLHL35 | LAMTOR2 | 0.747694 | 3 | 0 | 3 |
| KLHL35 | TIGD5 | 0.74591 | 3 | 0 | 3 |
| KLHL35 | AGAP2 | 0.744064 | 3 | 0 | 3 |
| KLHL35 | SPEF1 | 0.741815 | 3 | 0 | 3 |
| KLHL35 | RNF123 | 0.738754 | 4 | 0 | 3 |
| KLHL35 | CHIT1 | 0.728874 | 3 | 0 | 3 |
| KLHL35 | ZNF81 | 0.720984 | 3 | 0 | 3 |
| KLHL35 | SHANK1 | 0.718419 | 4 | 0 | 3 |
| KLHL35 | C1QL1 | 0.71548 | 3 | 0 | 3 |
| KLHL35 | SEMA3F | 0.713432 | 3 | 0 | 3 |
| KLHL35 | CHRNB4 | 0.712054 | 3 | 0 | 3 |
| KLHL35 | PRKCG | 0.711222 | 3 | 0 | 3 |
| KLHL35 | GCK | 0.711039 | 3 | 0 | 3 |
| KLHL35 | KRT35 | 0.710637 | 4 | 0 | 3 |
| KLHL35 | FOXB1 | 0.709469 | 4 | 0 | 4 |
For details and further investigation, click here