| Full name: ATPase H+ transporting V0 subunit e2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7q36.1 | ||
| Entrez ID: 155066 | HGNC ID: HGNC:21723 | Ensembl Gene: ENSG00000171130 | OMIM ID: 611019 |
| Drug and gene relationship at DGIdb | |||
ATP6V0E2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection |
Expression of ATP6V0E2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V0E2 | 155066 | 213587_s_at | 0.0456 | 0.9724 | |
| GSE20347 | ATP6V0E2 | 155066 | 213587_s_at | 0.5569 | 0.0167 | |
| GSE23400 | ATP6V0E2 | 155066 | 213587_s_at | -0.1586 | 0.2275 | |
| GSE26886 | ATP6V0E2 | 155066 | 213587_s_at | 0.2035 | 0.5802 | |
| GSE29001 | ATP6V0E2 | 155066 | 213587_s_at | -0.0888 | 0.8666 | |
| GSE38129 | ATP6V0E2 | 155066 | 213587_s_at | 0.0826 | 0.7574 | |
| GSE45670 | ATP6V0E2 | 155066 | 213587_s_at | -0.4530 | 0.1557 | |
| GSE53622 | ATP6V0E2 | 155066 | 9952 | 0.0091 | 0.9010 | |
| GSE53624 | ATP6V0E2 | 155066 | 9952 | -0.0256 | 0.7214 | |
| GSE63941 | ATP6V0E2 | 155066 | 213587_s_at | -1.0318 | 0.2055 | |
| GSE77861 | ATP6V0E2 | 155066 | 213587_s_at | -0.2425 | 0.2923 | |
| GSE97050 | ATP6V0E2 | 155066 | A_23_P61960 | -0.2991 | 0.6558 | |
| SRP007169 | ATP6V0E2 | 155066 | RNAseq | -1.2379 | 0.0788 | |
| SRP064894 | ATP6V0E2 | 155066 | RNAseq | -0.3684 | 0.4327 | |
| SRP133303 | ATP6V0E2 | 155066 | RNAseq | -0.3857 | 0.3756 | |
| SRP159526 | ATP6V0E2 | 155066 | RNAseq | 0.5110 | 0.1894 | |
| SRP193095 | ATP6V0E2 | 155066 | RNAseq | 0.0920 | 0.7141 | |
| SRP219564 | ATP6V0E2 | 155066 | RNAseq | -0.6425 | 0.3180 | |
| TCGA | ATP6V0E2 | 155066 | RNAseq | -0.5315 | 0.0048 |
Upregulated datasets: 0; Downregulated datasets: 0.
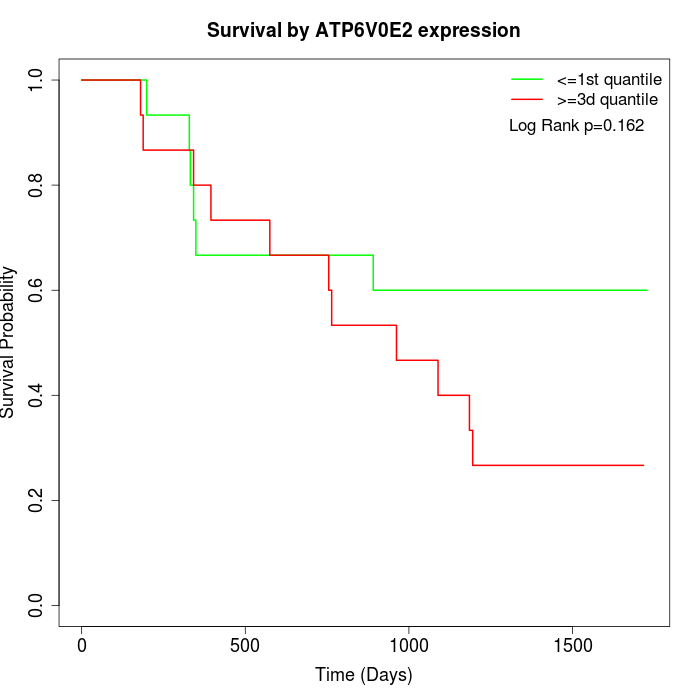
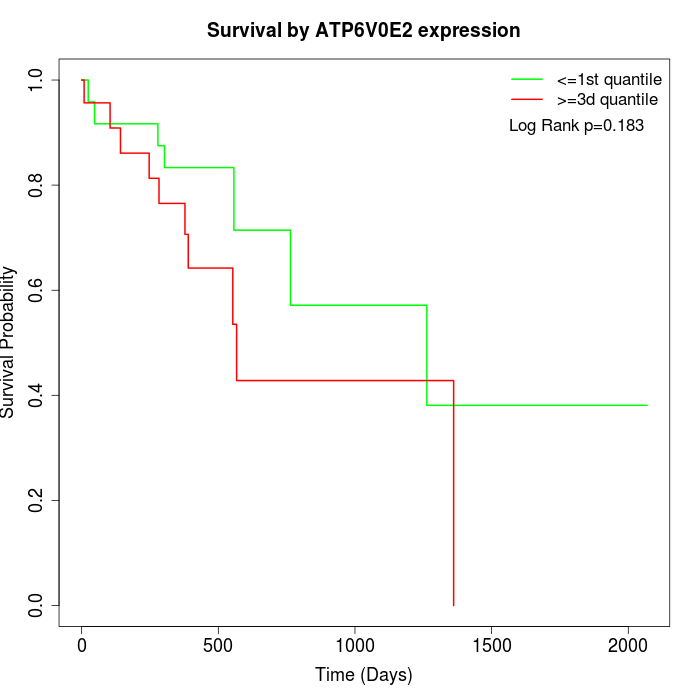
Survival by ATP6V0E2 expression:
Note: Click image to view full size file.
Copy number change of ATP6V0E2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V0E2 | 155066 | 2 | 3 | 25 | |
| GSE20123 | ATP6V0E2 | 155066 | 2 | 3 | 25 | |
| GSE43470 | ATP6V0E2 | 155066 | 2 | 5 | 36 | |
| GSE46452 | ATP6V0E2 | 155066 | 7 | 2 | 50 | |
| GSE47630 | ATP6V0E2 | 155066 | 6 | 8 | 26 | |
| GSE54993 | ATP6V0E2 | 155066 | 3 | 3 | 64 | |
| GSE54994 | ATP6V0E2 | 155066 | 6 | 8 | 39 | |
| GSE60625 | ATP6V0E2 | 155066 | 0 | 0 | 11 | |
| GSE74703 | ATP6V0E2 | 155066 | 2 | 4 | 30 | |
| GSE74704 | ATP6V0E2 | 155066 | 1 | 3 | 16 | |
| TCGA | ATP6V0E2 | 155066 | 25 | 27 | 44 |
Total number of gains: 56; Total number of losses: 66; Total Number of normals: 366.
Somatic mutations of ATP6V0E2:
Generating mutation plots.
Highly correlated genes for ATP6V0E2:
Showing top 20/324 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V0E2 | SPSB4 | 0.736899 | 3 | 0 | 3 |
| ATP6V0E2 | GSTT1 | 0.730068 | 3 | 0 | 3 |
| ATP6V0E2 | CCNB3 | 0.728631 | 4 | 0 | 4 |
| ATP6V0E2 | KRTAP19-2 | 0.72776 | 3 | 0 | 3 |
| ATP6V0E2 | GDPD1 | 0.714211 | 3 | 0 | 3 |
| ATP6V0E2 | ZNF546 | 0.698156 | 3 | 0 | 3 |
| ATP6V0E2 | SFTPA2 | 0.695821 | 3 | 0 | 3 |
| ATP6V0E2 | ABHD16A | 0.694179 | 3 | 0 | 3 |
| ATP6V0E2 | FOXP4 | 0.691265 | 5 | 0 | 5 |
| ATP6V0E2 | MB | 0.688523 | 3 | 0 | 3 |
| ATP6V0E2 | ZNF225 | 0.688052 | 5 | 0 | 4 |
| ATP6V0E2 | HP1BP3 | 0.68796 | 3 | 0 | 3 |
| ATP6V0E2 | HLCS | 0.685287 | 3 | 0 | 3 |
| ATP6V0E2 | ZNF784 | 0.683209 | 4 | 0 | 3 |
| ATP6V0E2 | ZNF136 | 0.674818 | 3 | 0 | 3 |
| ATP6V0E2 | NDUFB11 | 0.672462 | 4 | 0 | 4 |
| ATP6V0E2 | DYM | 0.671638 | 3 | 0 | 3 |
| ATP6V0E2 | PYGO1 | 0.670647 | 4 | 0 | 4 |
| ATP6V0E2 | THAP9 | 0.667014 | 3 | 0 | 3 |
| ATP6V0E2 | TTC33 | 0.664223 | 4 | 0 | 4 |
For details and further investigation, click here