| Full name: myoglobin | Alias Symbol: PVALB | ||
| Type: protein-coding gene | Cytoband: 22q12.3 | ||
| Entrez ID: 4151 | HGNC ID: HGNC:6915 | Ensembl Gene: ENSG00000198125 | OMIM ID: 160000 |
| Drug and gene relationship at DGIdb | |||
Expression of MB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MB | 4151 | 204179_at | 0.3154 | 0.7971 | |
| GSE20347 | MB | 4151 | 204179_at | 0.0766 | 0.6602 | |
| GSE23400 | MB | 4151 | 204179_at | -0.0941 | 0.2711 | |
| GSE26886 | MB | 4151 | 204179_at | 0.3883 | 0.2966 | |
| GSE29001 | MB | 4151 | 204179_at | -0.0358 | 0.8665 | |
| GSE38129 | MB | 4151 | 204179_at | -0.0708 | 0.8013 | |
| GSE45670 | MB | 4151 | 204179_at | 0.6081 | 0.0960 | |
| GSE53622 | MB | 4151 | 21737 | -0.1847 | 0.2295 | |
| GSE53624 | MB | 4151 | 21737 | -0.1911 | 0.0624 | |
| GSE63941 | MB | 4151 | 204179_at | 1.9065 | 0.1662 | |
| GSE77861 | MB | 4151 | 204179_at | 0.0271 | 0.8293 | |
| GSE97050 | MB | 4151 | A_33_P3303649 | -1.8888 | 0.1249 | |
| SRP219564 | MB | 4151 | RNAseq | -1.4475 | 0.1200 | |
| TCGA | MB | 4151 | RNAseq | 0.0588 | 0.9154 |
Upregulated datasets: 0; Downregulated datasets: 0.
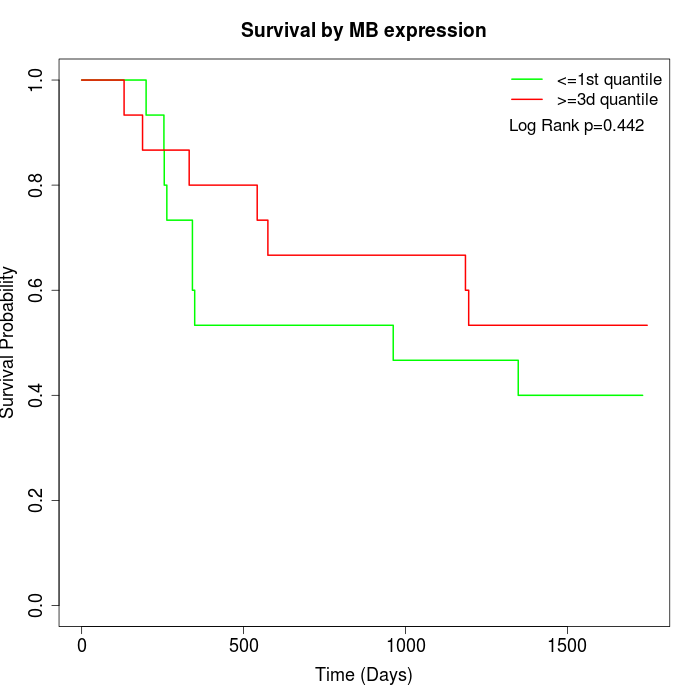
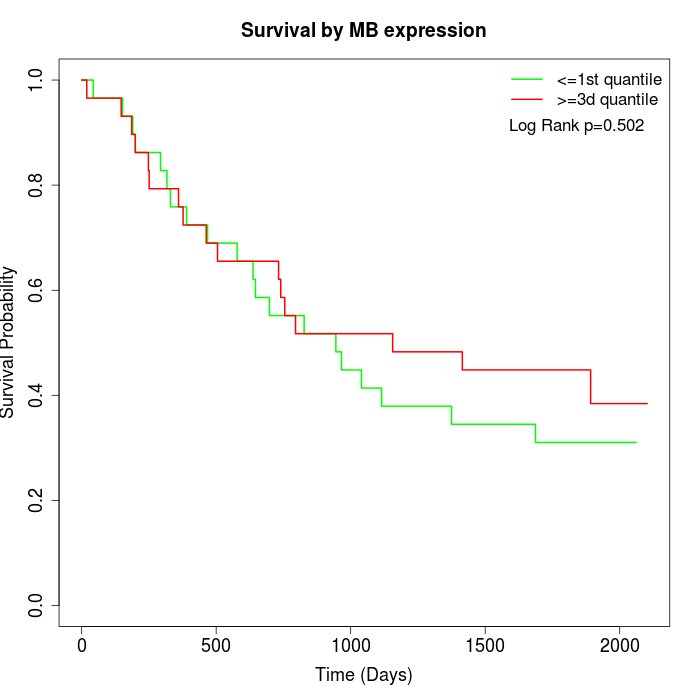
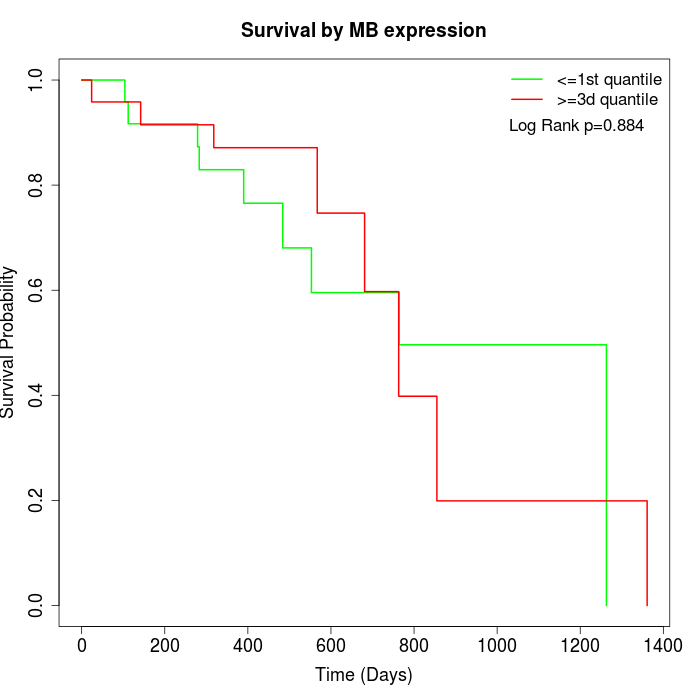
Survival by MB expression:
Note: Click image to view full size file.
Copy number change of MB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MB | 4151 | 4 | 6 | 20 | |
| GSE20123 | MB | 4151 | 4 | 5 | 21 | |
| GSE43470 | MB | 4151 | 4 | 7 | 32 | |
| GSE46452 | MB | 4151 | 31 | 1 | 27 | |
| GSE47630 | MB | 4151 | 8 | 5 | 27 | |
| GSE54993 | MB | 4151 | 3 | 6 | 61 | |
| GSE54994 | MB | 4151 | 9 | 9 | 35 | |
| GSE60625 | MB | 4151 | 5 | 0 | 6 | |
| GSE74703 | MB | 4151 | 4 | 5 | 27 | |
| GSE74704 | MB | 4151 | 1 | 2 | 17 | |
| TCGA | MB | 4151 | 27 | 15 | 54 |
Total number of gains: 100; Total number of losses: 61; Total Number of normals: 327.
Somatic mutations of MB:
Generating mutation plots.
Highly correlated genes for MB:
Showing top 20/216 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MB | SHANK2 | 0.714438 | 3 | 0 | 3 |
| MB | FAM43B | 0.711061 | 3 | 0 | 3 |
| MB | NELL1 | 0.699229 | 3 | 0 | 3 |
| MB | KLHL24 | 0.699006 | 4 | 0 | 3 |
| MB | MYL2 | 0.695498 | 3 | 0 | 3 |
| MB | PCSK2 | 0.689382 | 4 | 0 | 4 |
| MB | ATP6V0E2 | 0.688523 | 3 | 0 | 3 |
| MB | MYBPC1 | 0.682844 | 5 | 0 | 4 |
| MB | CTXN1 | 0.682384 | 3 | 0 | 3 |
| MB | TUB | 0.677321 | 3 | 0 | 3 |
| MB | RNF150 | 0.668977 | 3 | 0 | 3 |
| MB | MYL1 | 0.661926 | 5 | 0 | 3 |
| MB | PPP1R9A | 0.651098 | 4 | 0 | 3 |
| MB | CSRP3 | 0.650756 | 4 | 0 | 3 |
| MB | DCP1B | 0.650647 | 3 | 0 | 3 |
| MB | FN3K | 0.649552 | 4 | 0 | 3 |
| MB | SLC2A4 | 0.636311 | 3 | 0 | 3 |
| MB | CDX1 | 0.636074 | 4 | 0 | 4 |
| MB | DCHS2 | 0.633711 | 6 | 0 | 4 |
| MB | CNTN2 | 0.633687 | 3 | 0 | 3 |
For details and further investigation, click here