| Full name: splA/ryanodine receptor domain and SOCS box containing 4 | Alias Symbol: SSB-4 | ||
| Type: protein-coding gene | Cytoband: 3q23 | ||
| Entrez ID: 92369 | HGNC ID: HGNC:30630 | Ensembl Gene: ENSG00000175093 | OMIM ID: 611660 |
| Drug and gene relationship at DGIdb | |||
Expression of SPSB4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SPSB4 | 92369 | 229929_at | 0.2023 | 0.4011 | |
| GSE26886 | SPSB4 | 92369 | 229929_at | 0.0753 | 0.6626 | |
| GSE45670 | SPSB4 | 92369 | 229929_at | -0.0116 | 0.9186 | |
| GSE53622 | SPSB4 | 92369 | 133424 | 0.0690 | 0.4438 | |
| GSE53624 | SPSB4 | 92369 | 133424 | -0.0926 | 0.3622 | |
| GSE63941 | SPSB4 | 92369 | 229929_at | 0.2398 | 0.2956 | |
| GSE77861 | SPSB4 | 92369 | 229929_at | -0.0391 | 0.8004 | |
| GSE97050 | SPSB4 | 92369 | A_23_P258381 | -0.0919 | 0.6792 | |
| SRP159526 | SPSB4 | 92369 | RNAseq | 5.1762 | 0.0000 | |
| SRP219564 | SPSB4 | 92369 | RNAseq | 1.2255 | 0.3439 | |
| TCGA | SPSB4 | 92369 | RNAseq | 0.7067 | 0.0804 |
Upregulated datasets: 1; Downregulated datasets: 0.
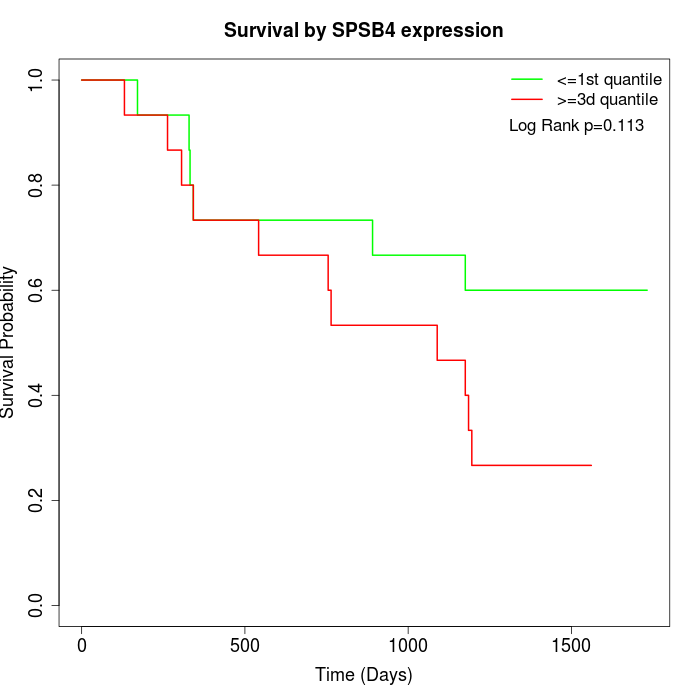
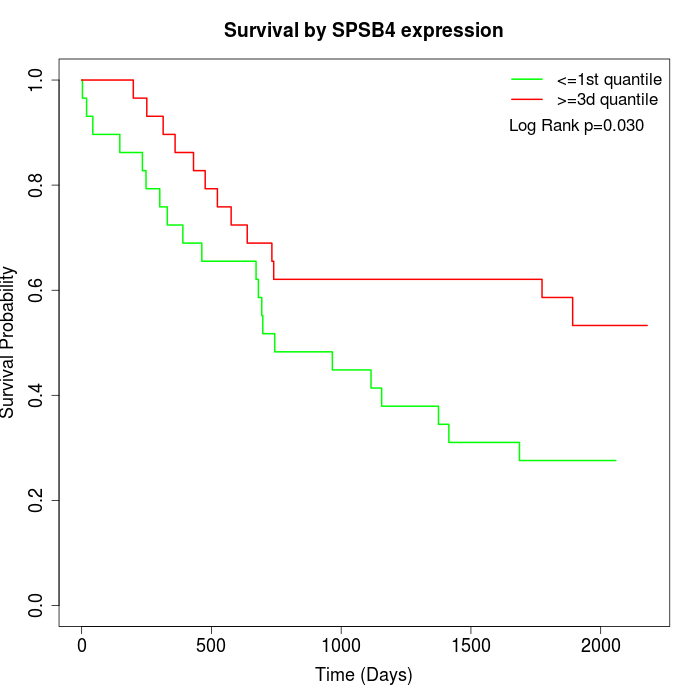
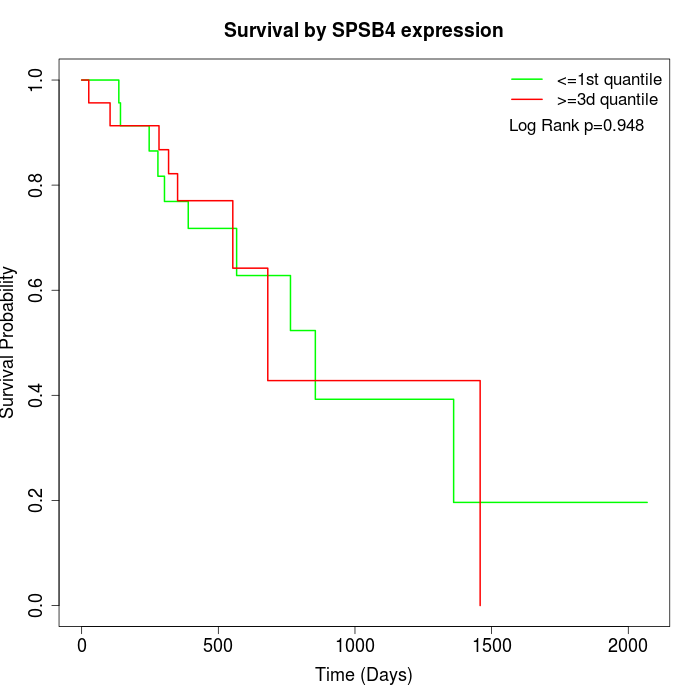
Survival by SPSB4 expression:
Note: Click image to view full size file.
Copy number change of SPSB4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SPSB4 | 92369 | 19 | 0 | 11 | |
| GSE20123 | SPSB4 | 92369 | 19 | 0 | 11 | |
| GSE43470 | SPSB4 | 92369 | 22 | 0 | 21 | |
| GSE46452 | SPSB4 | 92369 | 15 | 4 | 40 | |
| GSE47630 | SPSB4 | 92369 | 19 | 3 | 18 | |
| GSE54993 | SPSB4 | 92369 | 1 | 8 | 61 | |
| GSE54994 | SPSB4 | 92369 | 34 | 1 | 18 | |
| GSE60625 | SPSB4 | 92369 | 0 | 6 | 5 | |
| GSE74703 | SPSB4 | 92369 | 19 | 0 | 17 | |
| GSE74704 | SPSB4 | 92369 | 13 | 0 | 7 | |
| TCGA | SPSB4 | 92369 | 62 | 4 | 30 |
Total number of gains: 223; Total number of losses: 26; Total Number of normals: 239.
Somatic mutations of SPSB4:
Generating mutation plots.
Highly correlated genes for SPSB4:
Showing top 20/255 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SPSB4 | SFTPA2 | 0.894583 | 3 | 0 | 3 |
| SPSB4 | CYP26C1 | 0.792592 | 3 | 0 | 3 |
| SPSB4 | TMEM150A | 0.784757 | 3 | 0 | 3 |
| SPSB4 | CTPS2 | 0.780846 | 3 | 0 | 3 |
| SPSB4 | KRTAP21-2 | 0.780124 | 3 | 0 | 3 |
| SPSB4 | KIR3DL2 | 0.773663 | 3 | 0 | 3 |
| SPSB4 | NFKBID | 0.756834 | 3 | 0 | 3 |
| SPSB4 | ARTN | 0.746747 | 4 | 0 | 4 |
| SPSB4 | MAPRE3 | 0.742197 | 3 | 0 | 3 |
| SPSB4 | ATP6V0E2 | 0.736899 | 3 | 0 | 3 |
| SPSB4 | OR2T33 | 0.73184 | 3 | 0 | 3 |
| SPSB4 | IP6K1 | 0.729686 | 3 | 0 | 3 |
| SPSB4 | KRTAP19-2 | 0.728722 | 3 | 0 | 3 |
| SPSB4 | NEIL2 | 0.727307 | 3 | 0 | 3 |
| SPSB4 | THRAP3 | 0.725482 | 3 | 0 | 3 |
| SPSB4 | ENTPD2 | 0.724809 | 5 | 0 | 5 |
| SPSB4 | KRTAP9-6 | 0.72432 | 3 | 0 | 3 |
| SPSB4 | KRTAP10-9 | 0.723262 | 3 | 0 | 3 |
| SPSB4 | ZNF225 | 0.720446 | 3 | 0 | 3 |
| SPSB4 | LRRC36 | 0.714941 | 4 | 0 | 4 |
For details and further investigation, click here