| Full name: pygopus family PHD finger 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 15q21.3 | ||
| Entrez ID: 26108 | HGNC ID: HGNC:30256 | Ensembl Gene: ENSG00000171016 | OMIM ID: 606902 |
| Drug and gene relationship at DGIdb | |||
Expression of PYGO1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PYGO1 | 26108 | 239741_at | 0.0238 | 0.9867 | |
| GSE20347 | PYGO1 | 26108 | 215517_at | -0.1415 | 0.0152 | |
| GSE23400 | PYGO1 | 26108 | 215517_at | -0.1467 | 0.0000 | |
| GSE26886 | PYGO1 | 26108 | 215517_at | 0.1813 | 0.1553 | |
| GSE29001 | PYGO1 | 26108 | 215517_at | 0.0155 | 0.9596 | |
| GSE38129 | PYGO1 | 26108 | 215517_at | -0.1422 | 0.0643 | |
| GSE45670 | PYGO1 | 26108 | 215517_at | -0.3356 | 0.0035 | |
| GSE53622 | PYGO1 | 26108 | 12594 | 0.0821 | 0.6815 | |
| GSE53624 | PYGO1 | 26108 | 12594 | 0.2319 | 0.1419 | |
| GSE63941 | PYGO1 | 26108 | 215517_at | -0.6312 | 0.1619 | |
| GSE77861 | PYGO1 | 26108 | 215517_at | -0.1148 | 0.4225 | |
| GSE97050 | PYGO1 | 26108 | A_32_P530933 | -0.4617 | 0.2462 | |
| SRP007169 | PYGO1 | 26108 | RNAseq | 1.5886 | 0.0256 | |
| SRP008496 | PYGO1 | 26108 | RNAseq | 2.3813 | 0.0000 | |
| SRP064894 | PYGO1 | 26108 | RNAseq | 0.1254 | 0.6779 | |
| SRP133303 | PYGO1 | 26108 | RNAseq | 0.4011 | 0.1022 | |
| SRP159526 | PYGO1 | 26108 | RNAseq | 0.9100 | 0.0004 | |
| SRP193095 | PYGO1 | 26108 | RNAseq | 0.4469 | 0.0144 | |
| SRP219564 | PYGO1 | 26108 | RNAseq | 0.2471 | 0.6372 | |
| TCGA | PYGO1 | 26108 | RNAseq | -0.2097 | 0.5019 |
Upregulated datasets: 2; Downregulated datasets: 0.
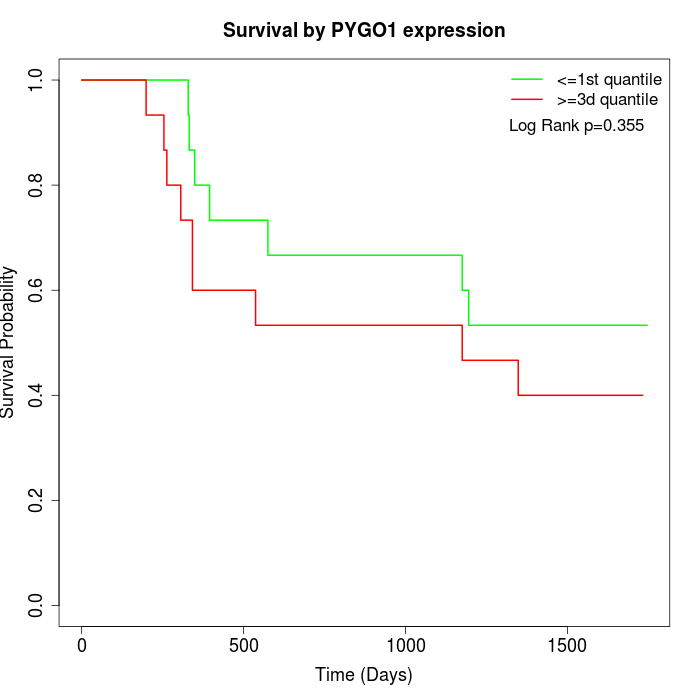
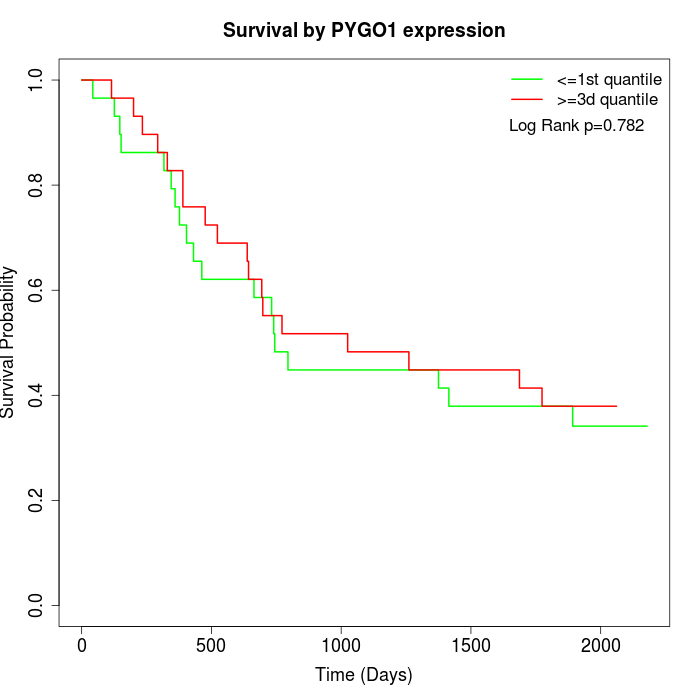
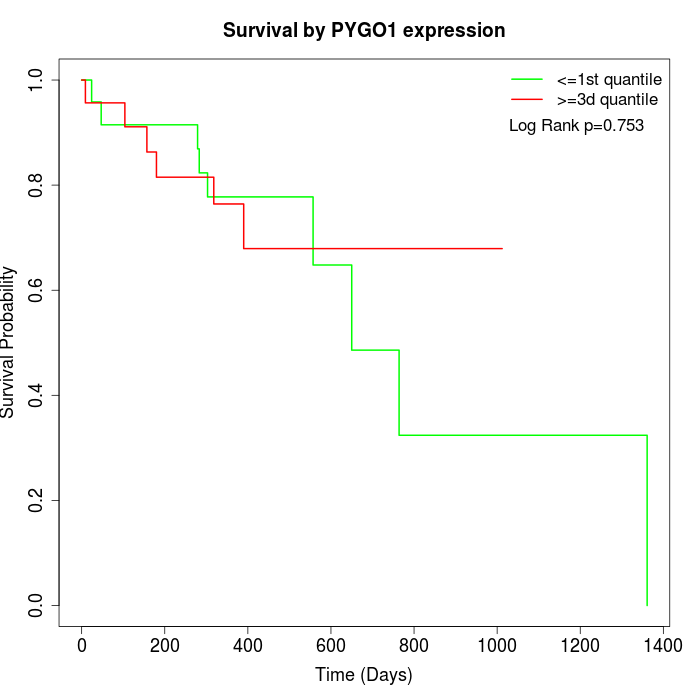
Survival by PYGO1 expression:
Note: Click image to view full size file.
Copy number change of PYGO1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PYGO1 | 26108 | 6 | 3 | 21 | |
| GSE20123 | PYGO1 | 26108 | 6 | 3 | 21 | |
| GSE43470 | PYGO1 | 26108 | 4 | 3 | 36 | |
| GSE46452 | PYGO1 | 26108 | 3 | 7 | 49 | |
| GSE47630 | PYGO1 | 26108 | 8 | 10 | 22 | |
| GSE54993 | PYGO1 | 26108 | 5 | 6 | 59 | |
| GSE54994 | PYGO1 | 26108 | 5 | 8 | 40 | |
| GSE60625 | PYGO1 | 26108 | 4 | 0 | 7 | |
| GSE74703 | PYGO1 | 26108 | 4 | 2 | 30 | |
| GSE74704 | PYGO1 | 26108 | 2 | 3 | 15 | |
| TCGA | PYGO1 | 26108 | 10 | 16 | 70 |
Total number of gains: 57; Total number of losses: 61; Total Number of normals: 370.
Somatic mutations of PYGO1:
Generating mutation plots.
Highly correlated genes for PYGO1:
Showing top 20/402 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PYGO1 | FAM98B | 0.753288 | 3 | 0 | 3 |
| PYGO1 | LRTOMT | 0.731183 | 3 | 0 | 3 |
| PYGO1 | GPR3 | 0.730736 | 3 | 0 | 3 |
| PYGO1 | CADM3 | 0.720854 | 3 | 0 | 3 |
| PYGO1 | IFNAR1 | 0.720808 | 3 | 0 | 3 |
| PYGO1 | PRR14 | 0.715146 | 3 | 0 | 3 |
| PYGO1 | GNAS | 0.708218 | 3 | 0 | 3 |
| PYGO1 | PIAS2 | 0.69853 | 3 | 0 | 3 |
| PYGO1 | ABHD14B | 0.698366 | 4 | 0 | 4 |
| PYGO1 | SH2B1 | 0.694958 | 4 | 0 | 4 |
| PYGO1 | CTPS2 | 0.693086 | 4 | 0 | 3 |
| PYGO1 | LNP1 | 0.693005 | 4 | 0 | 3 |
| PYGO1 | NCAM1 | 0.684352 | 4 | 0 | 4 |
| PYGO1 | DCAKD | 0.682458 | 3 | 0 | 3 |
| PYGO1 | COMMD1 | 0.680637 | 3 | 0 | 3 |
| PYGO1 | CDKL5 | 0.680075 | 5 | 0 | 5 |
| PYGO1 | TADA3 | 0.679572 | 3 | 0 | 3 |
| PYGO1 | TBC1D25 | 0.675575 | 3 | 0 | 3 |
| PYGO1 | RHAG | 0.673728 | 3 | 0 | 3 |
| PYGO1 | TSPAN18 | 0.67342 | 4 | 0 | 3 |
For details and further investigation, click here