| Full name: C-C motif chemokine receptor 10 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 17q21.2 | ||
| Entrez ID: 2826 | HGNC ID: HGNC:4474 | Ensembl Gene: ENSG00000184451 | OMIM ID: 600240 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CCR10 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway |
Expression of CCR10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCR10 | 2826 | 220565_at | -0.0357 | 0.9564 | |
| GSE20347 | CCR10 | 2826 | 220565_at | -0.0756 | 0.2049 | |
| GSE23400 | CCR10 | 2826 | 220565_at | -0.0398 | 0.1484 | |
| GSE26886 | CCR10 | 2826 | 220565_at | 0.5130 | 0.0554 | |
| GSE29001 | CCR10 | 2826 | 220565_at | -0.0692 | 0.6722 | |
| GSE38129 | CCR10 | 2826 | 220565_at | -0.1164 | 0.5419 | |
| GSE45670 | CCR10 | 2826 | 220565_at | -0.4181 | 0.0001 | |
| GSE53622 | CCR10 | 2826 | 16587 | 0.0205 | 0.8701 | |
| GSE53624 | CCR10 | 2826 | 16587 | -0.3843 | 0.0003 | |
| GSE63941 | CCR10 | 2826 | 220565_at | -0.1815 | 0.2611 | |
| GSE77861 | CCR10 | 2826 | 220565_at | -0.0702 | 0.7010 | |
| GSE97050 | CCR10 | 2826 | A_33_P3221303 | 0.1457 | 0.7737 | |
| SRP133303 | CCR10 | 2826 | RNAseq | 0.0372 | 0.9182 | |
| SRP159526 | CCR10 | 2826 | RNAseq | 0.7138 | 0.2570 | |
| SRP219564 | CCR10 | 2826 | RNAseq | -0.2075 | 0.7947 | |
| TCGA | CCR10 | 2826 | RNAseq | -0.6165 | 0.0149 |
Upregulated datasets: 0; Downregulated datasets: 0.
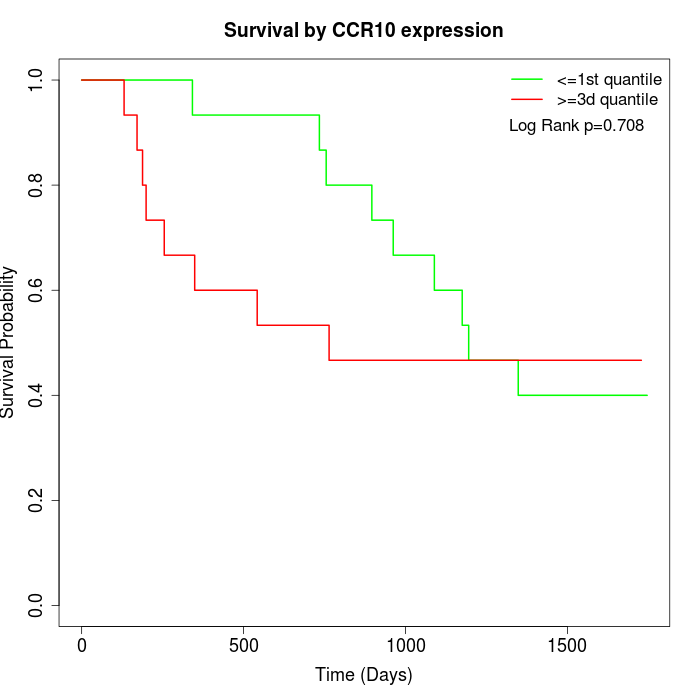
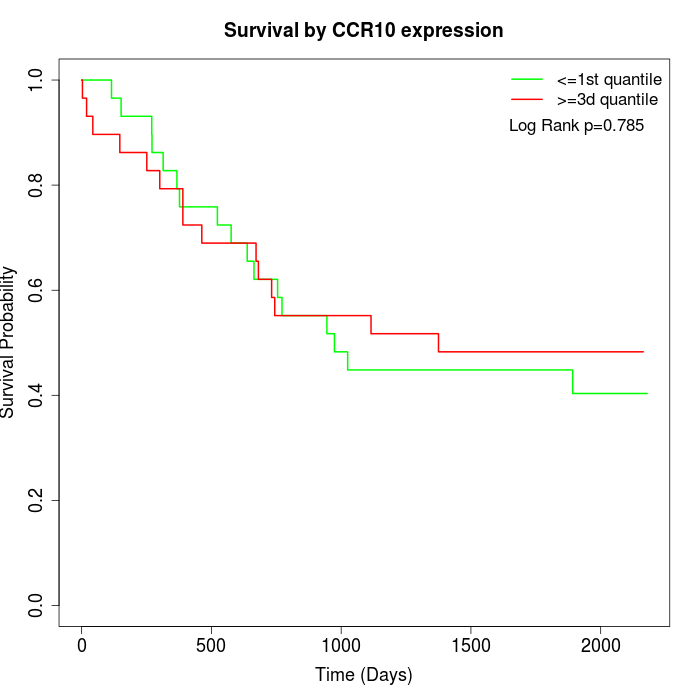
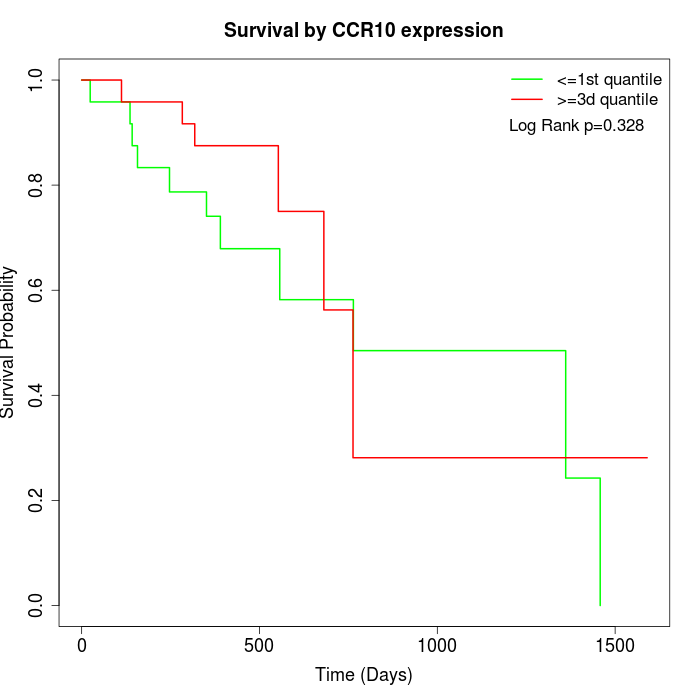
Survival by CCR10 expression:
Note: Click image to view full size file.
Copy number change of CCR10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCR10 | 2826 | 6 | 2 | 22 | |
| GSE20123 | CCR10 | 2826 | 6 | 2 | 22 | |
| GSE43470 | CCR10 | 2826 | 1 | 2 | 40 | |
| GSE46452 | CCR10 | 2826 | 34 | 0 | 25 | |
| GSE47630 | CCR10 | 2826 | 8 | 1 | 31 | |
| GSE54993 | CCR10 | 2826 | 3 | 4 | 63 | |
| GSE54994 | CCR10 | 2826 | 8 | 5 | 40 | |
| GSE60625 | CCR10 | 2826 | 4 | 0 | 7 | |
| GSE74703 | CCR10 | 2826 | 1 | 1 | 34 | |
| GSE74704 | CCR10 | 2826 | 4 | 1 | 15 | |
| TCGA | CCR10 | 2826 | 23 | 6 | 67 |
Total number of gains: 98; Total number of losses: 24; Total Number of normals: 366.
Somatic mutations of CCR10:
Generating mutation plots.
Highly correlated genes for CCR10:
Showing top 20/518 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCR10 | RBP2 | 0.777746 | 3 | 0 | 3 |
| CCR10 | FAM167B | 0.768553 | 3 | 0 | 3 |
| CCR10 | RAB39B | 0.759968 | 4 | 0 | 3 |
| CCR10 | ZDHHC1 | 0.734828 | 3 | 0 | 3 |
| CCR10 | GSK3A | 0.729117 | 4 | 0 | 4 |
| CCR10 | AFMID | 0.727033 | 4 | 0 | 4 |
| CCR10 | TCAP | 0.718792 | 5 | 0 | 5 |
| CCR10 | CPT1C | 0.716805 | 4 | 0 | 3 |
| CCR10 | MAGEH1 | 0.714846 | 4 | 0 | 4 |
| CCR10 | MZF1 | 0.710986 | 3 | 0 | 3 |
| CCR10 | C9orf50 | 0.710626 | 4 | 0 | 4 |
| CCR10 | TPP1 | 0.707323 | 3 | 0 | 3 |
| CCR10 | ZNF747 | 0.704926 | 3 | 0 | 3 |
| CCR10 | NUDT16 | 0.699437 | 4 | 0 | 4 |
| CCR10 | IGK | 0.696307 | 4 | 0 | 4 |
| CCR10 | CTF1 | 0.696151 | 5 | 0 | 3 |
| CCR10 | MAPRE3 | 0.692806 | 5 | 0 | 4 |
| CCR10 | ELAVL4 | 0.691424 | 3 | 0 | 3 |
| CCR10 | GIMAP1 | 0.689129 | 4 | 0 | 3 |
| CCR10 | PIM2 | 0.683793 | 4 | 0 | 4 |
For details and further investigation, click here