| Full name: Pim-2 proto-oncogene, serine/threonine kinase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xp11.23 | ||
| Entrez ID: 11040 | HGNC ID: HGNC:8987 | Ensembl Gene: ENSG00000102096 | OMIM ID: 300295 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PIM2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05221 | Acute myeloid leukemia |
Expression of PIM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIM2 | 11040 | 204269_at | 0.3525 | 0.6223 | |
| GSE20347 | PIM2 | 11040 | 204269_at | 0.2631 | 0.0005 | |
| GSE23400 | PIM2 | 11040 | 204269_at | 0.1151 | 0.0491 | |
| GSE26886 | PIM2 | 11040 | 204269_at | 0.1871 | 0.1385 | |
| GSE29001 | PIM2 | 11040 | 204269_at | -0.0677 | 0.6412 | |
| GSE38129 | PIM2 | 11040 | 204269_at | 0.3580 | 0.0362 | |
| GSE45670 | PIM2 | 11040 | 204269_at | 0.3783 | 0.0988 | |
| GSE63941 | PIM2 | 11040 | 204269_at | 0.3795 | 0.1944 | |
| GSE77861 | PIM2 | 11040 | 204269_at | 0.2089 | 0.1206 | |
| GSE97050 | PIM2 | 11040 | A_24_P379104 | 1.1153 | 0.0880 | |
| SRP007169 | PIM2 | 11040 | RNAseq | 2.0114 | 0.0021 | |
| SRP008496 | PIM2 | 11040 | RNAseq | 1.6402 | 0.0003 | |
| SRP064894 | PIM2 | 11040 | RNAseq | 0.8896 | 0.0067 | |
| SRP133303 | PIM2 | 11040 | RNAseq | 0.8299 | 0.0336 | |
| SRP159526 | PIM2 | 11040 | RNAseq | 1.2603 | 0.0730 | |
| SRP193095 | PIM2 | 11040 | RNAseq | 0.6381 | 0.1059 | |
| SRP219564 | PIM2 | 11040 | RNAseq | 0.8867 | 0.0863 | |
| TCGA | PIM2 | 11040 | RNAseq | 0.1516 | 0.1806 |
Upregulated datasets: 2; Downregulated datasets: 0.
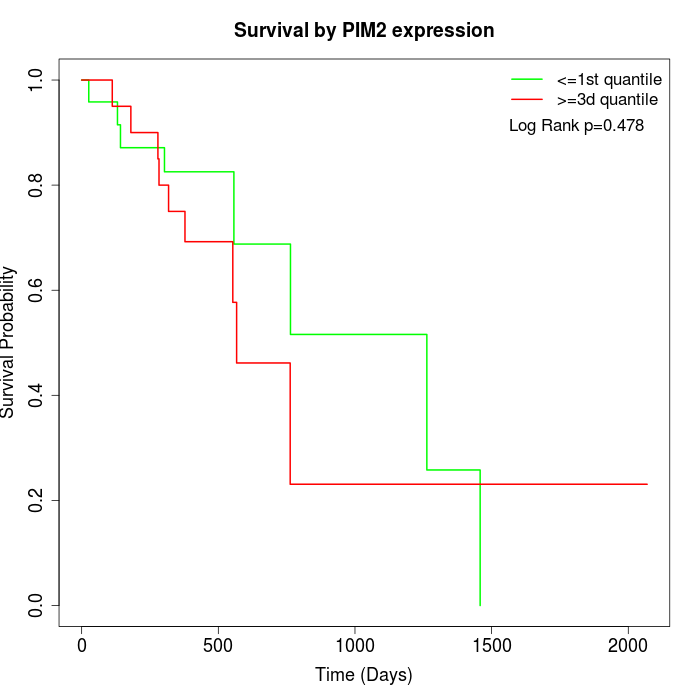
Survival by PIM2 expression:
Note: Click image to view full size file.
Copy number change of PIM2:
No record found for this gene.
Somatic mutations of PIM2:
Generating mutation plots.
Highly correlated genes for PIM2:
Showing top 20/430 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIM2 | TMEM214 | 0.771768 | 3 | 0 | 3 |
| PIM2 | BTLA | 0.752558 | 3 | 0 | 3 |
| PIM2 | CXCR5 | 0.750072 | 3 | 0 | 3 |
| PIM2 | ZBTB33 | 0.746864 | 3 | 0 | 3 |
| PIM2 | SMAP2 | 0.742199 | 4 | 0 | 4 |
| PIM2 | CTDSP2 | 0.740879 | 3 | 0 | 3 |
| PIM2 | SLAMF6 | 0.735044 | 3 | 0 | 3 |
| PIM2 | C5orf22 | 0.734638 | 3 | 0 | 3 |
| PIM2 | HGF | 0.726243 | 3 | 0 | 3 |
| PIM2 | MSC | 0.724535 | 3 | 0 | 3 |
| PIM2 | CD180 | 0.718531 | 6 | 0 | 6 |
| PIM2 | AKR1B1 | 0.713802 | 3 | 0 | 3 |
| PIM2 | VMA21 | 0.709312 | 3 | 0 | 3 |
| PIM2 | CCL22 | 0.707224 | 5 | 0 | 4 |
| PIM2 | MRM1 | 0.703418 | 3 | 0 | 3 |
| PIM2 | PARVG | 0.701071 | 4 | 0 | 4 |
| PIM2 | CNTNAP1 | 0.69683 | 4 | 0 | 4 |
| PIM2 | SEC11C | 0.695449 | 3 | 0 | 3 |
| PIM2 | HHEX | 0.694695 | 4 | 0 | 3 |
| PIM2 | IMPDH1 | 0.694423 | 3 | 0 | 3 |
For details and further investigation, click here