| Full name: egl-9 family hypoxia inducible factor 2 | Alias Symbol: PHD1|HIFPH1 | ||
| Type: protein-coding gene | Cytoband: 19q13.2 | ||
| Entrez ID: 112398 | HGNC ID: HGNC:14660 | Ensembl Gene: ENSG00000269858 | OMIM ID: 606424 |
| Drug and gene relationship at DGIdb | |||
EGLN2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04066 | HIF-1 signaling pathway | |
| hsa05200 | Pathways in cancer | |
| hsa05211 | Renal cell carcinoma |
Expression of EGLN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EGLN2 | 112398 | 227147_s_at | 0.1068 | 0.6175 | |
| GSE26886 | EGLN2 | 112398 | 227147_s_at | 0.1892 | 0.1181 | |
| GSE45670 | EGLN2 | 112398 | 227147_s_at | -0.0466 | 0.6932 | |
| GSE53622 | EGLN2 | 112398 | 98132 | -0.1959 | 0.0003 | |
| GSE53624 | EGLN2 | 112398 | 98132 | 0.2591 | 0.0000 | |
| GSE63941 | EGLN2 | 112398 | 227147_s_at | 0.0742 | 0.6462 | |
| GSE77861 | EGLN2 | 112398 | 227147_s_at | -0.1188 | 0.4262 | |
| GSE97050 | EGLN2 | 112398 | A_24_P322395 | -0.0936 | 0.6126 | |
| SRP007169 | EGLN2 | 112398 | RNAseq | -1.0154 | 0.0351 | |
| SRP008496 | EGLN2 | 112398 | RNAseq | -0.8224 | 0.0054 | |
| SRP064894 | EGLN2 | 112398 | RNAseq | 0.4674 | 0.1406 | |
| SRP133303 | EGLN2 | 112398 | RNAseq | 0.0376 | 0.8416 | |
| SRP159526 | EGLN2 | 112398 | RNAseq | 0.1510 | 0.6048 | |
| SRP193095 | EGLN2 | 112398 | RNAseq | -0.0958 | 0.4636 | |
| SRP219564 | EGLN2 | 112398 | RNAseq | -0.2366 | 0.4495 | |
| TCGA | EGLN2 | 112398 | RNAseq | -0.0763 | 0.1176 |
Upregulated datasets: 0; Downregulated datasets: 1.
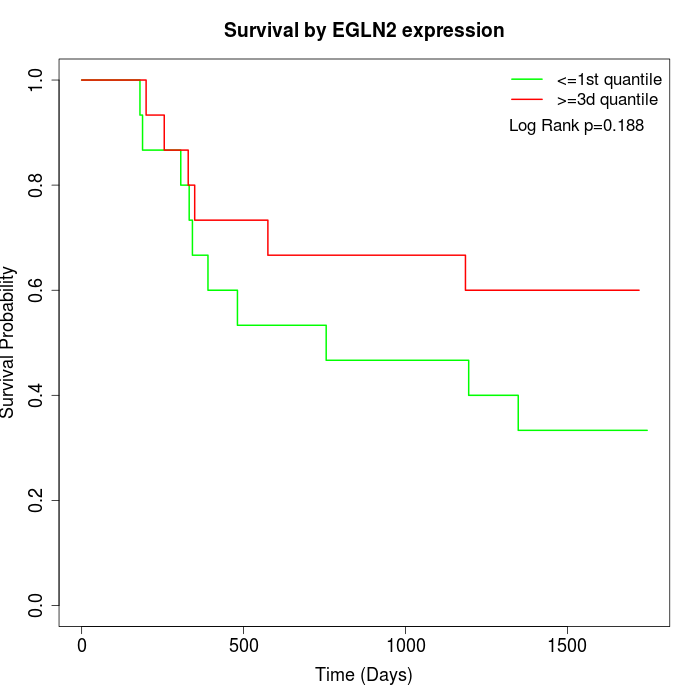
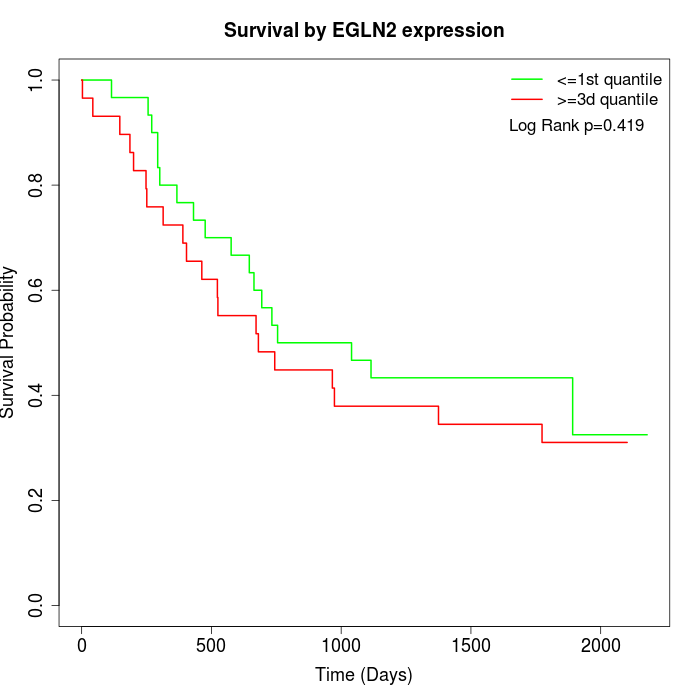
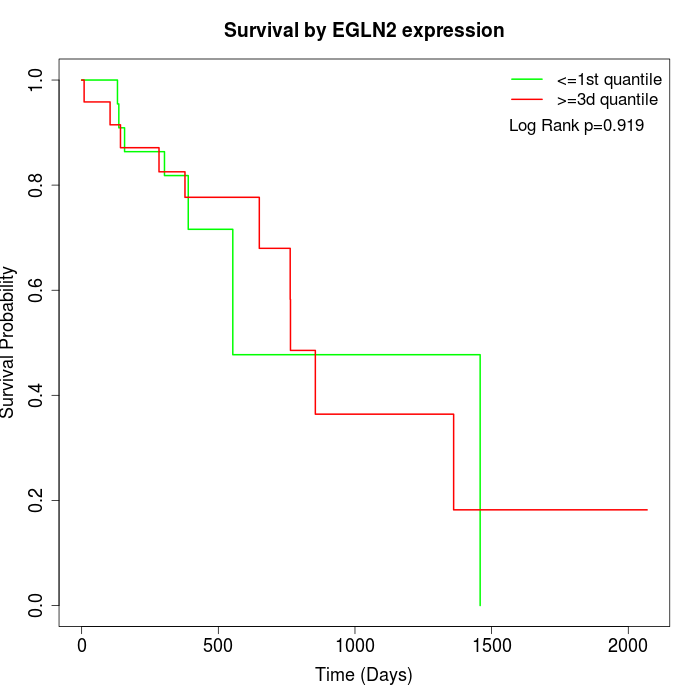
Survival by EGLN2 expression:
Note: Click image to view full size file.
Copy number change of EGLN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EGLN2 | 112398 | 4 | 5 | 21 | |
| GSE20123 | EGLN2 | 112398 | 4 | 4 | 22 | |
| GSE43470 | EGLN2 | 112398 | 3 | 9 | 31 | |
| GSE46452 | EGLN2 | 112398 | 48 | 1 | 10 | |
| GSE47630 | EGLN2 | 112398 | 9 | 5 | 26 | |
| GSE54993 | EGLN2 | 112398 | 17 | 4 | 49 | |
| GSE54994 | EGLN2 | 112398 | 8 | 9 | 36 | |
| GSE60625 | EGLN2 | 112398 | 9 | 0 | 2 | |
| GSE74703 | EGLN2 | 112398 | 3 | 6 | 27 | |
| GSE74704 | EGLN2 | 112398 | 4 | 2 | 14 | |
| TCGA | EGLN2 | 112398 | 14 | 16 | 66 |
Total number of gains: 123; Total number of losses: 61; Total Number of normals: 304.
Somatic mutations of EGLN2:
Generating mutation plots.
Highly correlated genes for EGLN2:
Showing top 20/153 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EGLN2 | PDE8A | 0.817382 | 3 | 0 | 3 |
| EGLN2 | SARDH | 0.790754 | 3 | 0 | 3 |
| EGLN2 | MAP1S | 0.772864 | 3 | 0 | 3 |
| EGLN2 | MILR1 | 0.771349 | 3 | 0 | 3 |
| EGLN2 | ABHD16B | 0.765235 | 4 | 0 | 4 |
| EGLN2 | UQCRFS1 | 0.759756 | 3 | 0 | 3 |
| EGLN2 | CDAN1 | 0.75016 | 3 | 0 | 3 |
| EGLN2 | JMJD4 | 0.748898 | 3 | 0 | 3 |
| EGLN2 | MAST1 | 0.738094 | 3 | 0 | 3 |
| EGLN2 | ANPEP | 0.736854 | 3 | 0 | 3 |
| EGLN2 | GNL3L | 0.728634 | 3 | 0 | 3 |
| EGLN2 | TBC1D16 | 0.724321 | 3 | 0 | 3 |
| EGLN2 | LIN37 | 0.720492 | 3 | 0 | 3 |
| EGLN2 | GAS2L1 | 0.718433 | 3 | 0 | 3 |
| EGLN2 | ATIC | 0.717244 | 3 | 0 | 3 |
| EGLN2 | NUBPL | 0.717122 | 3 | 0 | 3 |
| EGLN2 | EVI5 | 0.713527 | 3 | 0 | 3 |
| EGLN2 | PCBP3 | 0.713498 | 3 | 0 | 3 |
| EGLN2 | TMEM214 | 0.711406 | 3 | 0 | 3 |
| EGLN2 | ARL16 | 0.709088 | 3 | 0 | 3 |
For details and further investigation, click here