| Full name: microtubule associated protein 1S | Alias Symbol: FLJ10669|MAP8 | ||
| Type: protein-coding gene | Cytoband: 19p13.12 | ||
| Entrez ID: 55201 | HGNC ID: HGNC:15715 | Ensembl Gene: ENSG00000130479 | OMIM ID: 607573 |
| Drug and gene relationship at DGIdb | |||
Expression of MAP1S:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP1S | 55201 | 218522_s_at | 0.5198 | 0.3293 | |
| GSE20347 | MAP1S | 55201 | 218522_s_at | 0.1076 | 0.3328 | |
| GSE23400 | MAP1S | 55201 | 218522_s_at | -0.0216 | 0.7345 | |
| GSE26886 | MAP1S | 55201 | 218522_s_at | 0.5454 | 0.0077 | |
| GSE29001 | MAP1S | 55201 | 218522_s_at | -0.1640 | 0.5893 | |
| GSE38129 | MAP1S | 55201 | 218522_s_at | 0.1403 | 0.2624 | |
| GSE45670 | MAP1S | 55201 | 218522_s_at | -0.0140 | 0.9317 | |
| GSE53622 | MAP1S | 55201 | 46716 | 0.2827 | 0.0000 | |
| GSE53624 | MAP1S | 55201 | 46716 | 0.6905 | 0.0000 | |
| GSE63941 | MAP1S | 55201 | 218522_s_at | 0.2530 | 0.3929 | |
| GSE77861 | MAP1S | 55201 | 218522_s_at | -0.1467 | 0.4753 | |
| GSE97050 | MAP1S | 55201 | A_33_P3343090 | 0.1685 | 0.4347 | |
| SRP007169 | MAP1S | 55201 | RNAseq | -0.5202 | 0.3965 | |
| SRP008496 | MAP1S | 55201 | RNAseq | -0.7159 | 0.0757 | |
| SRP064894 | MAP1S | 55201 | RNAseq | 0.2877 | 0.2271 | |
| SRP133303 | MAP1S | 55201 | RNAseq | 0.2109 | 0.2219 | |
| SRP159526 | MAP1S | 55201 | RNAseq | 0.4135 | 0.1281 | |
| SRP193095 | MAP1S | 55201 | RNAseq | 0.2222 | 0.2386 | |
| SRP219564 | MAP1S | 55201 | RNAseq | 0.2191 | 0.5090 | |
| TCGA | MAP1S | 55201 | RNAseq | 0.0053 | 0.9216 |
Upregulated datasets: 0; Downregulated datasets: 0.
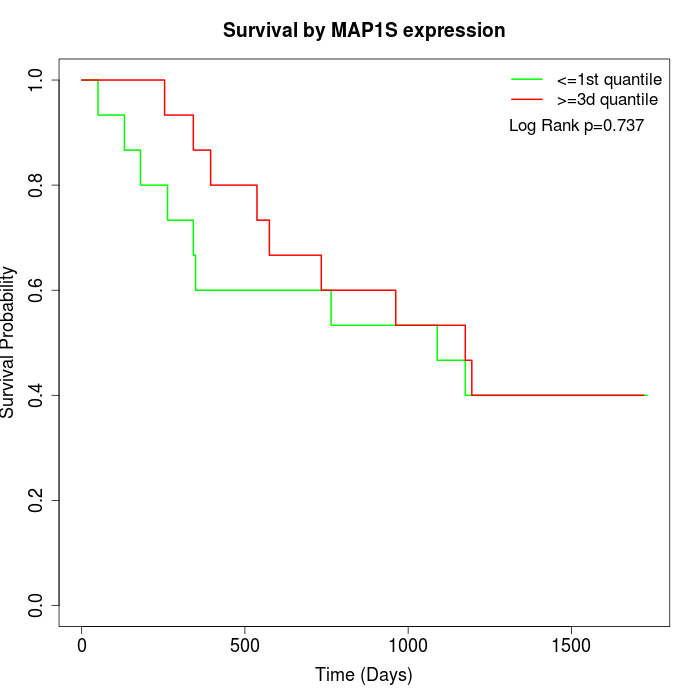
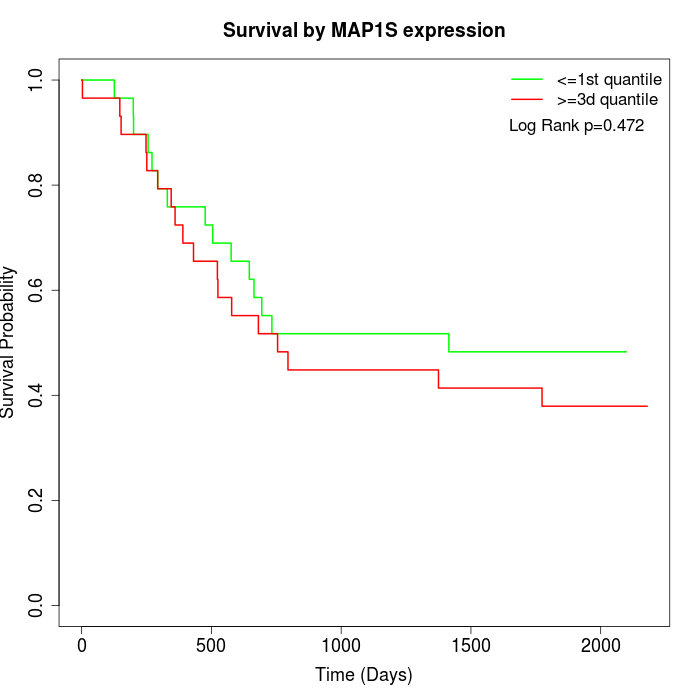
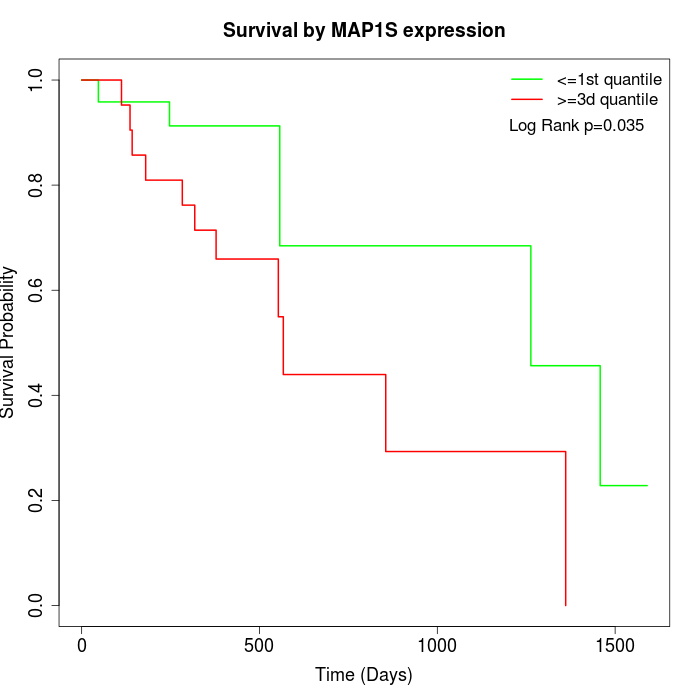
Survival by MAP1S expression:
Note: Click image to view full size file.
Copy number change of MAP1S:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP1S | 55201 | 4 | 4 | 22 | |
| GSE20123 | MAP1S | 55201 | 3 | 2 | 25 | |
| GSE43470 | MAP1S | 55201 | 2 | 6 | 35 | |
| GSE46452 | MAP1S | 55201 | 47 | 1 | 11 | |
| GSE47630 | MAP1S | 55201 | 4 | 8 | 28 | |
| GSE54993 | MAP1S | 55201 | 15 | 4 | 51 | |
| GSE54994 | MAP1S | 55201 | 6 | 14 | 33 | |
| GSE60625 | MAP1S | 55201 | 9 | 0 | 2 | |
| GSE74703 | MAP1S | 55201 | 2 | 4 | 30 | |
| GSE74704 | MAP1S | 55201 | 0 | 2 | 18 | |
| TCGA | MAP1S | 55201 | 17 | 9 | 70 |
Total number of gains: 109; Total number of losses: 54; Total Number of normals: 325.
Somatic mutations of MAP1S:
Generating mutation plots.
Highly correlated genes for MAP1S:
Showing top 20/408 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP1S | FOXP4 | 0.858233 | 3 | 0 | 3 |
| MAP1S | SLC26A11 | 0.79944 | 3 | 0 | 3 |
| MAP1S | NES | 0.789284 | 3 | 0 | 3 |
| MAP1S | WDTC1 | 0.786683 | 3 | 0 | 3 |
| MAP1S | EGLN2 | 0.772864 | 3 | 0 | 3 |
| MAP1S | CENPV | 0.771241 | 3 | 0 | 3 |
| MAP1S | AURKAIP1 | 0.767846 | 3 | 0 | 3 |
| MAP1S | TAS2R45 | 0.762241 | 3 | 0 | 3 |
| MAP1S | OPLAH | 0.745639 | 3 | 0 | 3 |
| MAP1S | ZMYM3 | 0.745169 | 3 | 0 | 3 |
| MAP1S | CCDC14 | 0.743139 | 3 | 0 | 3 |
| MAP1S | PLD6 | 0.738282 | 3 | 0 | 3 |
| MAP1S | ZFAND3 | 0.736621 | 3 | 0 | 3 |
| MAP1S | TAB1 | 0.734301 | 4 | 0 | 4 |
| MAP1S | GTDC1 | 0.732846 | 3 | 0 | 3 |
| MAP1S | DBP | 0.723115 | 3 | 0 | 3 |
| MAP1S | ARAF | 0.721284 | 3 | 0 | 3 |
| MAP1S | GTPBP6 | 0.717883 | 3 | 0 | 3 |
| MAP1S | ELFN1 | 0.715112 | 3 | 0 | 3 |
| MAP1S | YLPM1 | 0.710288 | 3 | 0 | 3 |
For details and further investigation, click here