| Full name: Fas apoptotic inhibitory molecule 2 | Alias Symbol: KIAA0950|LFG|NMP35|LIFEGUARD|TMBIM2|LFG2 | ||
| Type: protein-coding gene | Cytoband: 12q13.12 | ||
| Entrez ID: 23017 | HGNC ID: HGNC:17067 | Ensembl Gene: ENSG00000135472 | OMIM ID: 604306 |
| Drug and gene relationship at DGIdb | |||
Expression of FAIM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FAIM2 | 23017 | 203618_at | 0.0649 | 0.7388 | |
| GSE20347 | FAIM2 | 23017 | 203618_at | 0.0118 | 0.9011 | |
| GSE23400 | FAIM2 | 23017 | 203618_at | -0.1796 | 0.0000 | |
| GSE26886 | FAIM2 | 23017 | 203618_at | 0.2024 | 0.0739 | |
| GSE29001 | FAIM2 | 23017 | 203618_at | 0.0454 | 0.8062 | |
| GSE38129 | FAIM2 | 23017 | 203618_at | -0.1395 | 0.0468 | |
| GSE45670 | FAIM2 | 23017 | 203618_at | -0.0533 | 0.5695 | |
| GSE53622 | FAIM2 | 23017 | 67489 | -0.9277 | 0.0000 | |
| GSE53624 | FAIM2 | 23017 | 67489 | -1.1306 | 0.0000 | |
| GSE63941 | FAIM2 | 23017 | 203618_at | 0.1047 | 0.4085 | |
| GSE77861 | FAIM2 | 23017 | 203618_at | -0.0890 | 0.4759 | |
| GSE97050 | FAIM2 | 23017 | A_33_P3248982 | -0.2708 | 0.5313 | |
| SRP064894 | FAIM2 | 23017 | RNAseq | -1.0140 | 0.1106 | |
| SRP133303 | FAIM2 | 23017 | RNAseq | -2.8196 | 0.0000 | |
| SRP219564 | FAIM2 | 23017 | RNAseq | 0.3021 | 0.8367 | |
| TCGA | FAIM2 | 23017 | RNAseq | -1.8828 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 3.
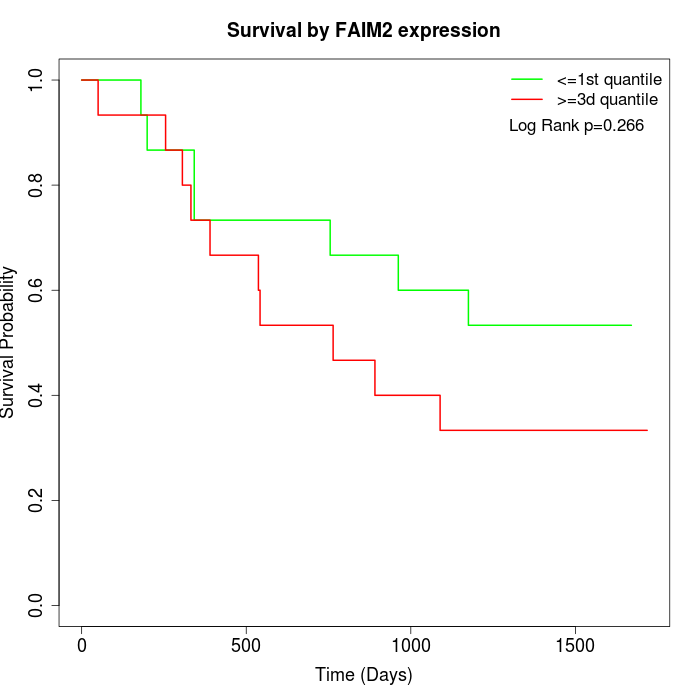
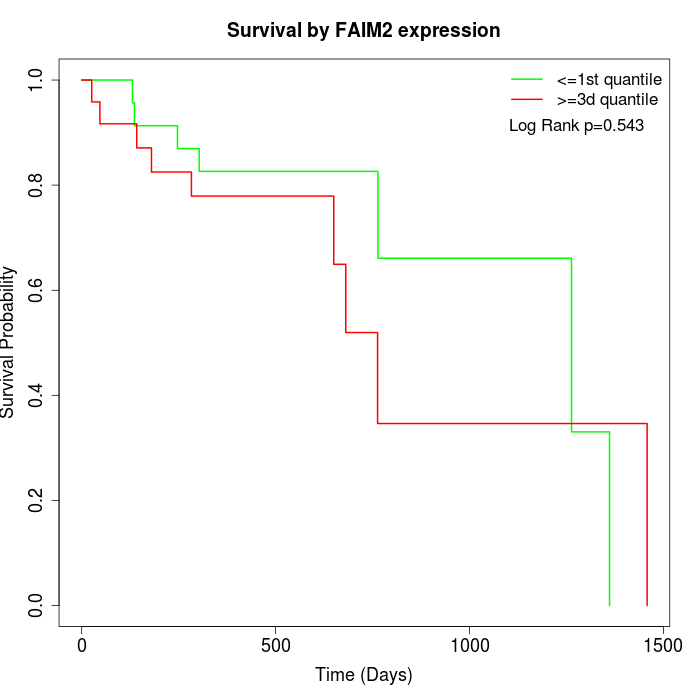
Survival by FAIM2 expression:
Note: Click image to view full size file.
Copy number change of FAIM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FAIM2 | 23017 | 7 | 1 | 22 | |
| GSE20123 | FAIM2 | 23017 | 7 | 1 | 22 | |
| GSE43470 | FAIM2 | 23017 | 3 | 0 | 40 | |
| GSE46452 | FAIM2 | 23017 | 8 | 1 | 50 | |
| GSE47630 | FAIM2 | 23017 | 11 | 2 | 27 | |
| GSE54993 | FAIM2 | 23017 | 0 | 5 | 65 | |
| GSE54994 | FAIM2 | 23017 | 4 | 1 | 48 | |
| GSE60625 | FAIM2 | 23017 | 0 | 0 | 11 | |
| GSE74703 | FAIM2 | 23017 | 3 | 0 | 33 | |
| GSE74704 | FAIM2 | 23017 | 5 | 1 | 14 | |
| TCGA | FAIM2 | 23017 | 15 | 11 | 70 |
Total number of gains: 63; Total number of losses: 23; Total Number of normals: 402.
Somatic mutations of FAIM2:
Generating mutation plots.
Highly correlated genes for FAIM2:
Showing top 20/735 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FAIM2 | BCAT2 | 0.75028 | 3 | 0 | 3 |
| FAIM2 | XKR7 | 0.739475 | 3 | 0 | 3 |
| FAIM2 | HLX | 0.733251 | 3 | 0 | 3 |
| FAIM2 | CABP7 | 0.717478 | 3 | 0 | 3 |
| FAIM2 | GPR26 | 0.70178 | 5 | 0 | 4 |
| FAIM2 | FBLN7 | 0.690736 | 3 | 0 | 3 |
| FAIM2 | RHBDL1 | 0.689031 | 3 | 0 | 3 |
| FAIM2 | TMEFF2 | 0.687757 | 3 | 0 | 3 |
| FAIM2 | MCEMP1 | 0.687043 | 3 | 0 | 3 |
| FAIM2 | MATN1 | 0.684198 | 3 | 0 | 3 |
| FAIM2 | NPHS1 | 0.678382 | 3 | 0 | 3 |
| FAIM2 | PALM3 | 0.677708 | 3 | 0 | 3 |
| FAIM2 | KCNF1 | 0.671932 | 7 | 0 | 6 |
| FAIM2 | CEND1 | 0.670823 | 7 | 0 | 6 |
| FAIM2 | MEF2C-AS1 | 0.669432 | 3 | 0 | 3 |
| FAIM2 | IL9R | 0.669222 | 6 | 0 | 6 |
| FAIM2 | C19orf81 | 0.668156 | 3 | 0 | 3 |
| FAIM2 | BTBD19 | 0.665765 | 5 | 0 | 4 |
| FAIM2 | PSG7 | 0.665145 | 3 | 0 | 3 |
| FAIM2 | ABCA4 | 0.663138 | 4 | 0 | 4 |
For details and further investigation, click here