| Full name: branched chain amino acid transaminase 2 | Alias Symbol: BCAM | ||
| Type: protein-coding gene | Cytoband: 19q13.33 | ||
| Entrez ID: 587 | HGNC ID: HGNC:977 | Ensembl Gene: ENSG00000105552 | OMIM ID: 113530 |
| Drug and gene relationship at DGIdb | |||
Expression of BCAT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BCAT2 | 587 | 203576_at | 0.5058 | 0.0908 | |
| GSE20347 | BCAT2 | 587 | 203576_at | 0.4224 | 0.0009 | |
| GSE23400 | BCAT2 | 587 | 203576_at | 0.1511 | 0.0211 | |
| GSE26886 | BCAT2 | 587 | 203576_at | 0.4180 | 0.0495 | |
| GSE29001 | BCAT2 | 587 | 215654_at | 0.0985 | 0.6150 | |
| GSE38129 | BCAT2 | 587 | 203576_at | 0.4815 | 0.0001 | |
| GSE45670 | BCAT2 | 587 | 203576_at | 0.1642 | 0.3422 | |
| GSE53622 | BCAT2 | 587 | 37086 | 0.0213 | 0.8276 | |
| GSE53624 | BCAT2 | 587 | 37086 | 0.1293 | 0.1101 | |
| GSE63941 | BCAT2 | 587 | 203576_at | 1.9585 | 0.0001 | |
| GSE77861 | BCAT2 | 587 | 203576_at | -0.0031 | 0.9915 | |
| GSE97050 | BCAT2 | 587 | A_23_P101551 | -0.1236 | 0.5795 | |
| SRP007169 | BCAT2 | 587 | RNAseq | 0.5537 | 0.1161 | |
| SRP008496 | BCAT2 | 587 | RNAseq | 0.2476 | 0.4482 | |
| SRP064894 | BCAT2 | 587 | RNAseq | 0.6825 | 0.0508 | |
| SRP133303 | BCAT2 | 587 | RNAseq | 0.3697 | 0.1485 | |
| SRP159526 | BCAT2 | 587 | RNAseq | 0.2617 | 0.3788 | |
| SRP193095 | BCAT2 | 587 | RNAseq | 0.2553 | 0.0369 | |
| SRP219564 | BCAT2 | 587 | RNAseq | -0.0291 | 0.9387 | |
| TCGA | BCAT2 | 587 | RNAseq | -0.2059 | 0.0004 |
Upregulated datasets: 1; Downregulated datasets: 0.
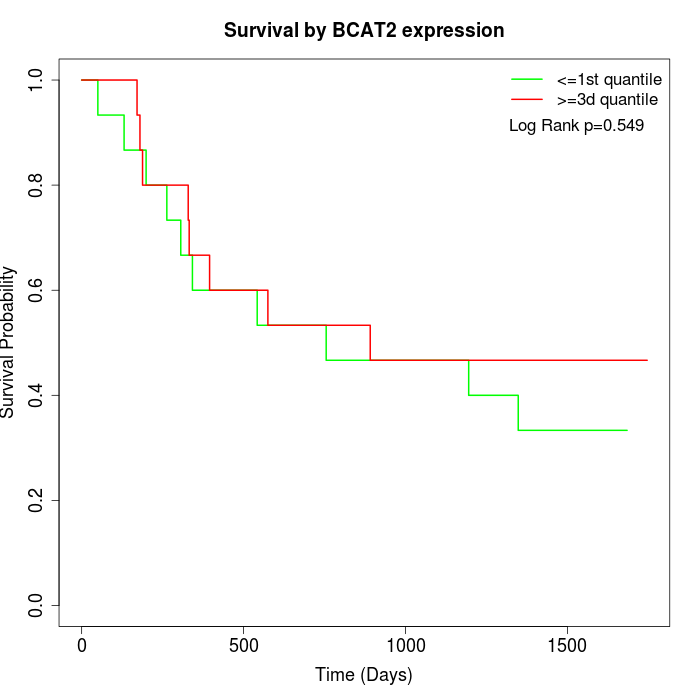
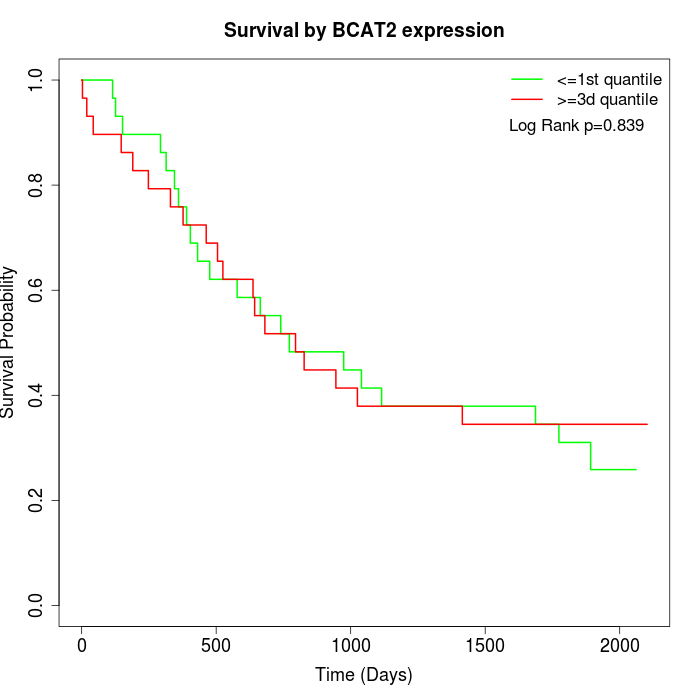
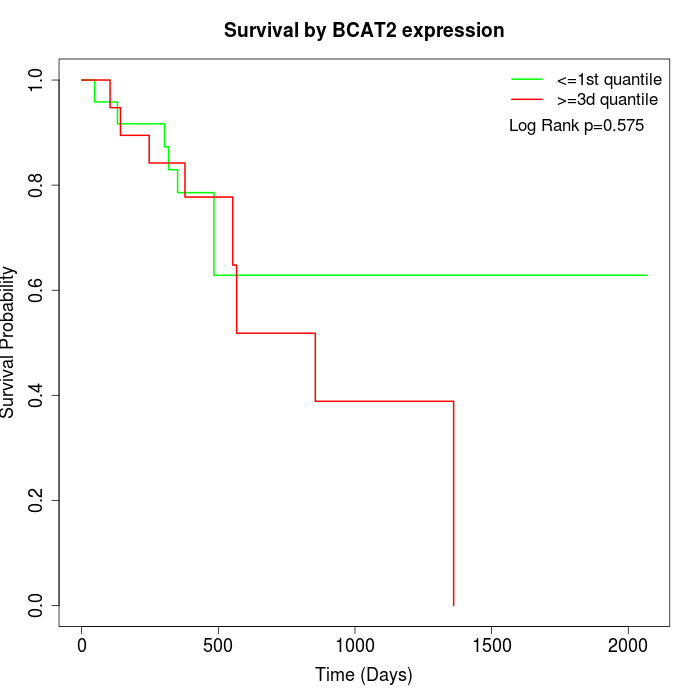
Survival by BCAT2 expression:
Note: Click image to view full size file.
Copy number change of BCAT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BCAT2 | 587 | 4 | 4 | 22 | |
| GSE20123 | BCAT2 | 587 | 4 | 3 | 23 | |
| GSE43470 | BCAT2 | 587 | 4 | 10 | 29 | |
| GSE46452 | BCAT2 | 587 | 45 | 1 | 13 | |
| GSE47630 | BCAT2 | 587 | 9 | 6 | 25 | |
| GSE54993 | BCAT2 | 587 | 17 | 4 | 49 | |
| GSE54994 | BCAT2 | 587 | 4 | 14 | 35 | |
| GSE60625 | BCAT2 | 587 | 9 | 0 | 2 | |
| GSE74703 | BCAT2 | 587 | 4 | 7 | 25 | |
| GSE74704 | BCAT2 | 587 | 4 | 1 | 15 | |
| TCGA | BCAT2 | 587 | 14 | 18 | 64 |
Total number of gains: 118; Total number of losses: 68; Total Number of normals: 302.
Somatic mutations of BCAT2:
Generating mutation plots.
Highly correlated genes for BCAT2:
Showing top 20/272 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BCAT2 | FRS3 | 0.792521 | 3 | 0 | 3 |
| BCAT2 | FAIM2 | 0.75028 | 3 | 0 | 3 |
| BCAT2 | TEAD2 | 0.720193 | 3 | 0 | 3 |
| BCAT2 | BCL2 | 0.709991 | 3 | 0 | 3 |
| BCAT2 | TRDN | 0.694275 | 3 | 0 | 3 |
| BCAT2 | CDH15 | 0.684554 | 3 | 0 | 3 |
| BCAT2 | SOGA1 | 0.68133 | 3 | 0 | 3 |
| BCAT2 | PDZD4 | 0.66132 | 3 | 0 | 3 |
| BCAT2 | ACVR2B | 0.653899 | 4 | 0 | 4 |
| BCAT2 | CACTIN | 0.651398 | 4 | 0 | 3 |
| BCAT2 | KDF1 | 0.649642 | 3 | 0 | 3 |
| BCAT2 | ZNF691 | 0.647973 | 5 | 0 | 5 |
| BCAT2 | HPS1 | 0.647892 | 3 | 0 | 3 |
| BCAT2 | DAXX | 0.64592 | 5 | 0 | 4 |
| BCAT2 | ATN1 | 0.640276 | 3 | 0 | 3 |
| BCAT2 | SAFB2 | 0.639261 | 3 | 0 | 3 |
| BCAT2 | PRAM1 | 0.63714 | 3 | 0 | 3 |
| BCAT2 | ACTR1B | 0.634832 | 4 | 0 | 3 |
| BCAT2 | COL9A1 | 0.634752 | 3 | 0 | 3 |
| BCAT2 | MAP2K7 | 0.629541 | 5 | 0 | 5 |
For details and further investigation, click here