| Full name: potassium voltage-gated channel modifier subfamily F member 1 | Alias Symbol: Kv5.1|kH1|IK8 | ||
| Type: protein-coding gene | Cytoband: 2p25.1 | ||
| Entrez ID: 3754 | HGNC ID: HGNC:6246 | Ensembl Gene: ENSG00000162975 | OMIM ID: 603787 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNF1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNF1 | 3754 | 210263_at | -0.2613 | 0.7024 | |
| GSE20347 | KCNF1 | 3754 | 210263_at | -0.1012 | 0.2298 | |
| GSE23400 | KCNF1 | 3754 | 210263_at | -0.2346 | 0.0000 | |
| GSE26886 | KCNF1 | 3754 | 210263_at | 0.2132 | 0.0632 | |
| GSE29001 | KCNF1 | 3754 | 210263_at | -0.0010 | 0.9941 | |
| GSE38129 | KCNF1 | 3754 | 210263_at | -0.2516 | 0.0481 | |
| GSE45670 | KCNF1 | 3754 | 210263_at | -0.0734 | 0.5366 | |
| GSE53622 | KCNF1 | 3754 | 123394 | 0.0196 | 0.8857 | |
| GSE53624 | KCNF1 | 3754 | 123394 | 0.3305 | 0.0013 | |
| GSE63941 | KCNF1 | 3754 | 210263_at | -0.1086 | 0.5880 | |
| GSE77861 | KCNF1 | 3754 | 210263_at | -0.2031 | 0.1810 | |
| GSE97050 | KCNF1 | 3754 | A_33_P3424062 | -0.4139 | 0.3955 | |
| TCGA | KCNF1 | 3754 | RNAseq | -0.0848 | 0.8492 |
Upregulated datasets: 0; Downregulated datasets: 0.
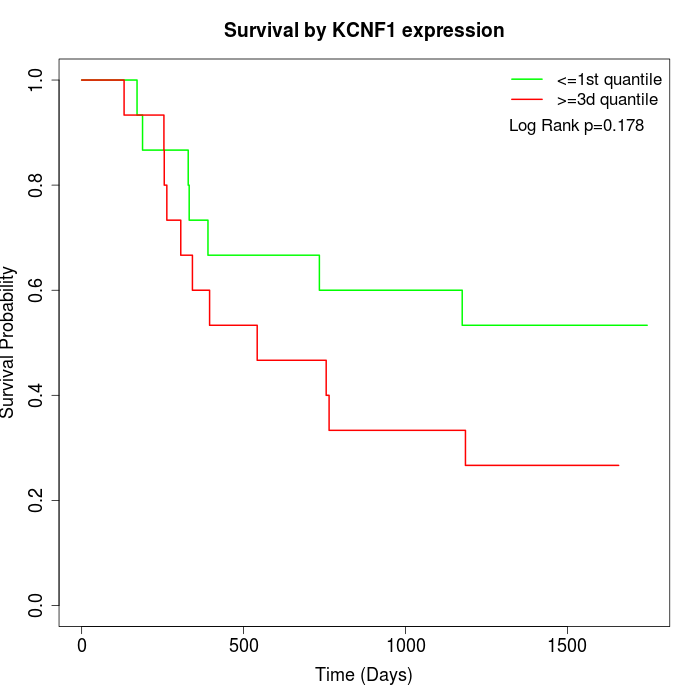
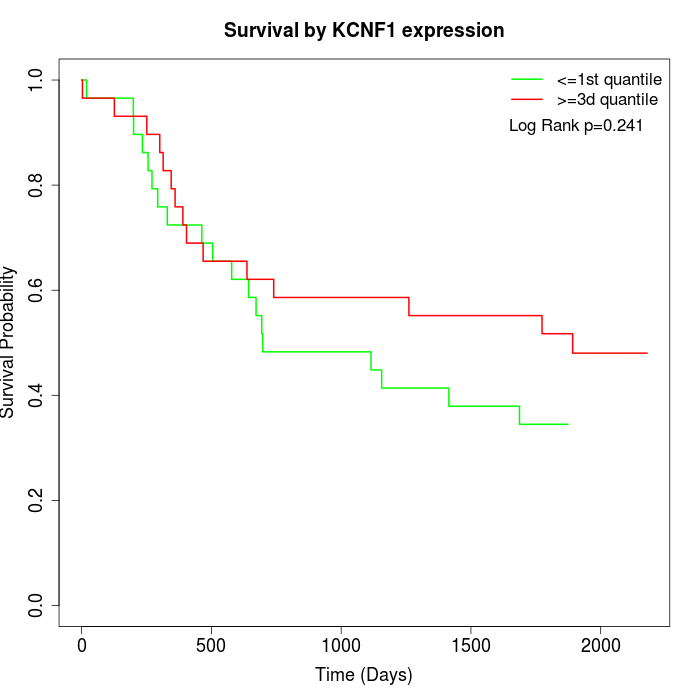
Survival by KCNF1 expression:
Note: Click image to view full size file.
Copy number change of KCNF1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNF1 | 3754 | 10 | 2 | 18 | |
| GSE20123 | KCNF1 | 3754 | 10 | 2 | 18 | |
| GSE43470 | KCNF1 | 3754 | 2 | 1 | 40 | |
| GSE46452 | KCNF1 | 3754 | 3 | 4 | 52 | |
| GSE47630 | KCNF1 | 3754 | 7 | 0 | 33 | |
| GSE54993 | KCNF1 | 3754 | 0 | 7 | 63 | |
| GSE54994 | KCNF1 | 3754 | 11 | 0 | 42 | |
| GSE60625 | KCNF1 | 3754 | 0 | 3 | 8 | |
| GSE74703 | KCNF1 | 3754 | 2 | 0 | 34 | |
| GSE74704 | KCNF1 | 3754 | 9 | 1 | 10 | |
| TCGA | KCNF1 | 3754 | 34 | 5 | 57 |
Total number of gains: 88; Total number of losses: 25; Total Number of normals: 375.
Somatic mutations of KCNF1:
Generating mutation plots.
Highly correlated genes for KCNF1:
Showing top 20/1100 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNF1 | DCDC1 | 0.805586 | 3 | 0 | 3 |
| KCNF1 | KLHL10 | 0.800386 | 3 | 0 | 3 |
| KCNF1 | SLC25A47 | 0.79854 | 3 | 0 | 3 |
| KCNF1 | MMP21 | 0.771713 | 3 | 0 | 3 |
| KCNF1 | TMEM235 | 0.768677 | 3 | 0 | 3 |
| KCNF1 | STRC | 0.768414 | 3 | 0 | 3 |
| KCNF1 | SCARF2 | 0.762409 | 3 | 0 | 3 |
| KCNF1 | C19orf71 | 0.760612 | 3 | 0 | 3 |
| KCNF1 | ISYNA1 | 0.75699 | 4 | 0 | 4 |
| KCNF1 | OR2T1 | 0.75283 | 3 | 0 | 3 |
| KCNF1 | PLA2G2C | 0.749301 | 3 | 0 | 3 |
| KCNF1 | NLRP6 | 0.747618 | 3 | 0 | 3 |
| KCNF1 | BSND | 0.74581 | 4 | 0 | 4 |
| KCNF1 | TRIML2 | 0.744397 | 3 | 0 | 3 |
| KCNF1 | BBC3 | 0.74104 | 3 | 0 | 3 |
| KCNF1 | SHISA8 | 0.740241 | 4 | 0 | 3 |
| KCNF1 | FAM71A | 0.733726 | 4 | 0 | 4 |
| KCNF1 | CDHR3 | 0.733569 | 4 | 0 | 3 |
| KCNF1 | CHST10 | 0.732652 | 4 | 0 | 4 |
| KCNF1 | TEF | 0.7284 | 4 | 0 | 4 |
For details and further investigation, click here