| Full name: G protein-coupled receptor 26 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q26.13 | ||
| Entrez ID: 2849 | HGNC ID: HGNC:4481 | Ensembl Gene: ENSG00000154478 | OMIM ID: 604847 |
| Drug and gene relationship at DGIdb | |||
Expression of GPR26:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GPR26 | 2849 | 1553865_a_at | 0.1493 | 0.5798 | |
| GSE26886 | GPR26 | 2849 | 244617_at | -0.1215 | 0.5255 | |
| GSE45670 | GPR26 | 2849 | 244617_at | -0.0350 | 0.7349 | |
| GSE53622 | GPR26 | 2849 | 1412 | -0.0349 | 0.7351 | |
| GSE53624 | GPR26 | 2849 | 1412 | 0.3175 | 0.0037 | |
| GSE63941 | GPR26 | 2849 | 244617_at | -0.0402 | 0.8034 | |
| GSE77861 | GPR26 | 2849 | 244617_at | -0.1026 | 0.5277 | |
| GSE97050 | GPR26 | 2849 | A_23_P415541 | -0.5005 | 0.3343 | |
| SRP133303 | GPR26 | 2849 | RNAseq | -0.6317 | 0.3614 | |
| TCGA | GPR26 | 2849 | RNAseq | -1.1786 | 0.1397 |
Upregulated datasets: 0; Downregulated datasets: 0.
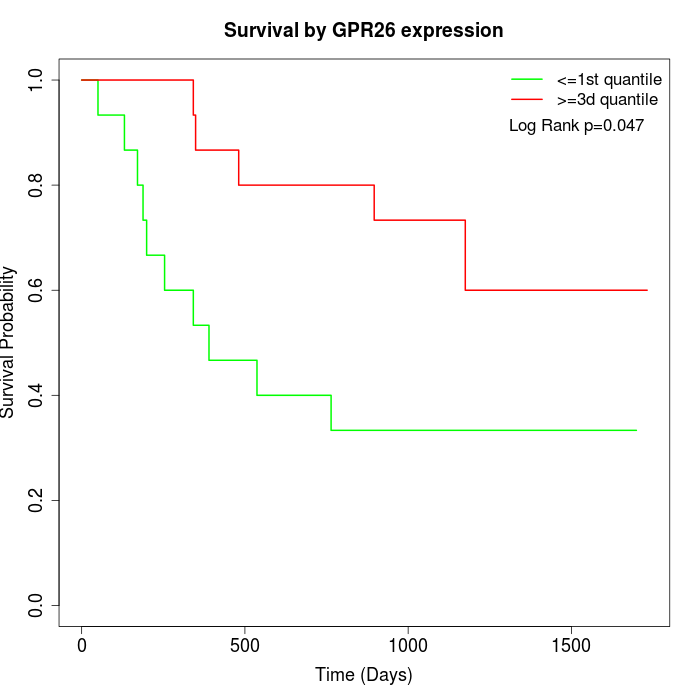
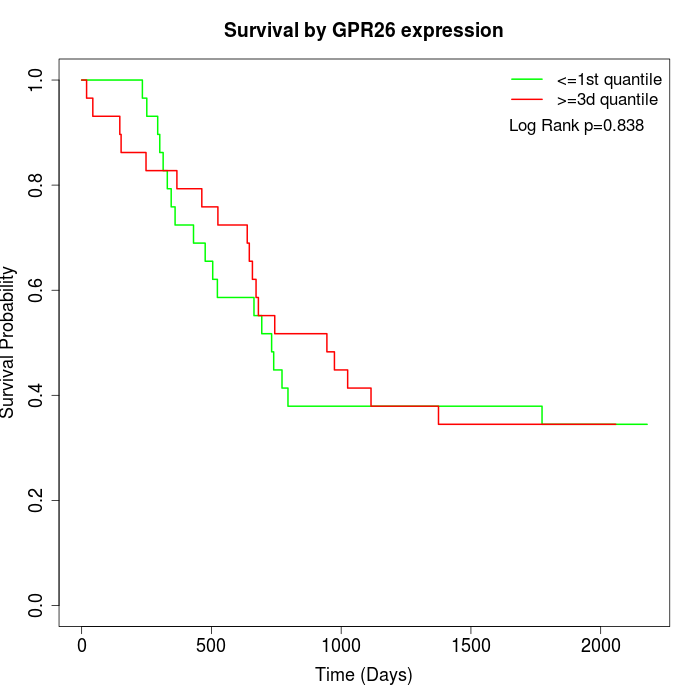
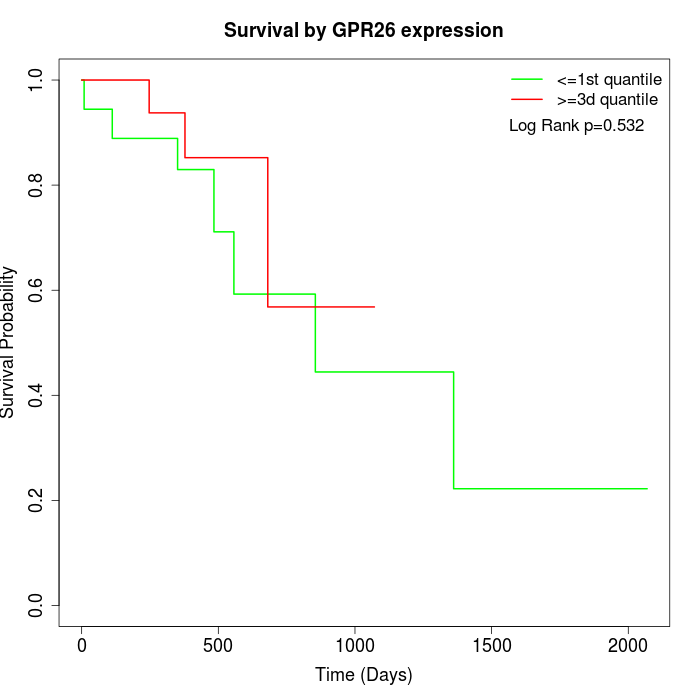
Survival by GPR26 expression:
Note: Click image to view full size file.
Copy number change of GPR26:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GPR26 | 2849 | 1 | 6 | 23 | |
| GSE20123 | GPR26 | 2849 | 1 | 6 | 23 | |
| GSE43470 | GPR26 | 2849 | 0 | 9 | 34 | |
| GSE46452 | GPR26 | 2849 | 0 | 11 | 48 | |
| GSE47630 | GPR26 | 2849 | 2 | 14 | 24 | |
| GSE54993 | GPR26 | 2849 | 8 | 1 | 61 | |
| GSE54994 | GPR26 | 2849 | 1 | 9 | 43 | |
| GSE60625 | GPR26 | 2849 | 0 | 0 | 11 | |
| GSE74703 | GPR26 | 2849 | 0 | 6 | 30 | |
| GSE74704 | GPR26 | 2849 | 1 | 2 | 17 | |
| TCGA | GPR26 | 2849 | 4 | 27 | 65 |
Total number of gains: 18; Total number of losses: 91; Total Number of normals: 379.
Somatic mutations of GPR26:
Generating mutation plots.
Highly correlated genes for GPR26:
Showing top 20/312 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GPR26 | EVX1 | 0.76094 | 3 | 0 | 3 |
| GPR26 | CFC1 | 0.736668 | 3 | 0 | 3 |
| GPR26 | MAG | 0.728154 | 3 | 0 | 3 |
| GPR26 | SCN2B | 0.711343 | 3 | 0 | 3 |
| GPR26 | FUT7 | 0.704028 | 3 | 0 | 3 |
| GPR26 | ADCY1 | 0.703796 | 3 | 0 | 3 |
| GPR26 | FAIM2 | 0.70178 | 5 | 0 | 4 |
| GPR26 | BCL2 | 0.701532 | 3 | 0 | 3 |
| GPR26 | IGH | 0.695113 | 3 | 0 | 3 |
| GPR26 | RASGRF2 | 0.693863 | 3 | 0 | 3 |
| GPR26 | FBXO44 | 0.691108 | 3 | 0 | 3 |
| GPR26 | DHRS7C | 0.690215 | 3 | 0 | 3 |
| GPR26 | PKLR | 0.689657 | 3 | 0 | 3 |
| GPR26 | SLC39A5 | 0.688415 | 4 | 0 | 3 |
| GPR26 | ATCAY | 0.6881 | 3 | 0 | 3 |
| GPR26 | TBKBP1 | 0.687739 | 3 | 0 | 3 |
| GPR26 | PEF1 | 0.687184 | 3 | 0 | 3 |
| GPR26 | DEFB124 | 0.68572 | 3 | 0 | 3 |
| GPR26 | PIH1D1 | 0.683079 | 3 | 0 | 3 |
| GPR26 | GALNT9 | 0.678652 | 4 | 0 | 4 |
For details and further investigation, click here