| Full name: X antigen family member 2 | Alias Symbol: XAGE-2|CT12.2 | ||
| Type: protein-coding gene | Cytoband: Xp11.22 | ||
| Entrez ID: 9502 | HGNC ID: HGNC:4112 | Ensembl Gene: ENSG00000155622 | OMIM ID: 300416 |
| Drug and gene relationship at DGIdb | |||
Expression of XAGE2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | XAGE2 | 9502 | 229469_at | -0.1212 | 0.5955 | |
| GSE26886 | XAGE2 | 9502 | 229469_at | 0.1310 | 0.1830 | |
| GSE45670 | XAGE2 | 9502 | 229469_at | -0.0328 | 0.7195 | |
| GSE53622 | XAGE2 | 9502 | 34432 | -0.0479 | 0.5892 | |
| GSE53624 | XAGE2 | 9502 | 34432 | 0.0432 | 0.6962 | |
| GSE63941 | XAGE2 | 9502 | 229469_at | 0.2133 | 0.2543 | |
| GSE77861 | XAGE2 | 9502 | 229469_at | -0.0615 | 0.6054 | |
| TCGA | XAGE2 | 9502 | RNAseq | 1.1708 | 0.6613 |
Upregulated datasets: 0; Downregulated datasets: 0.
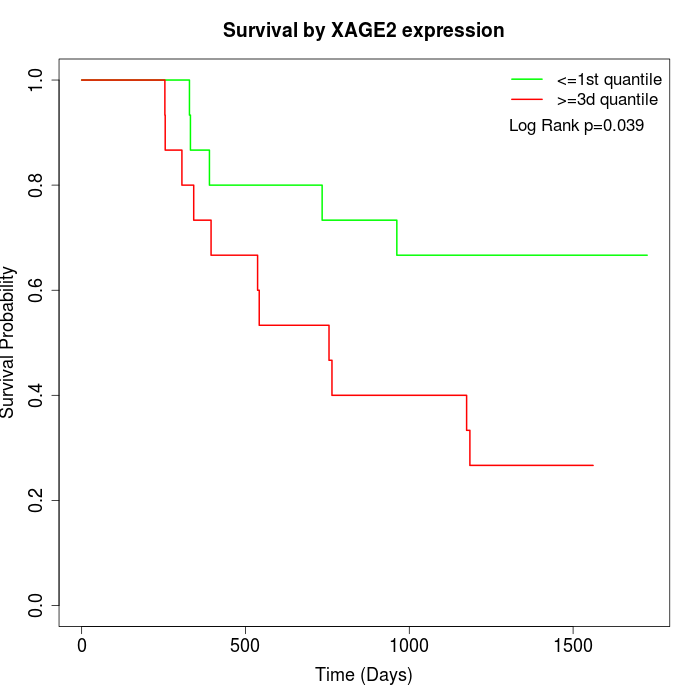
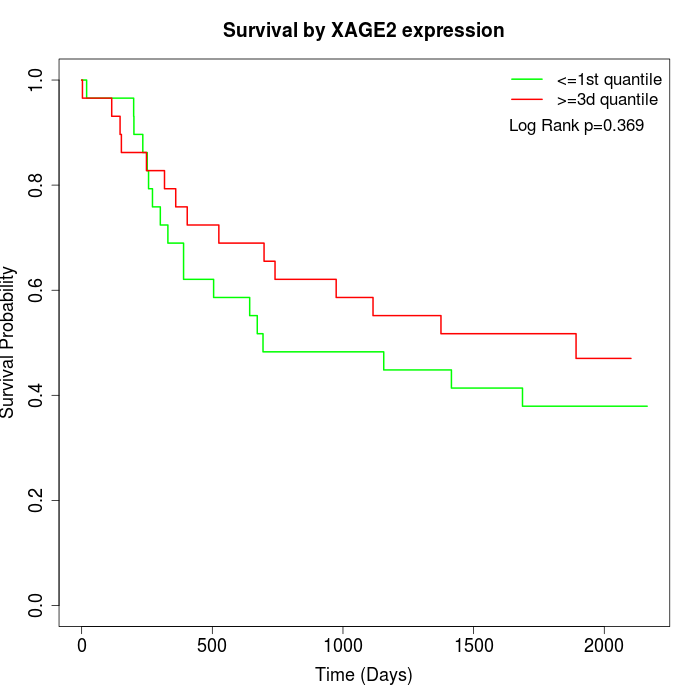
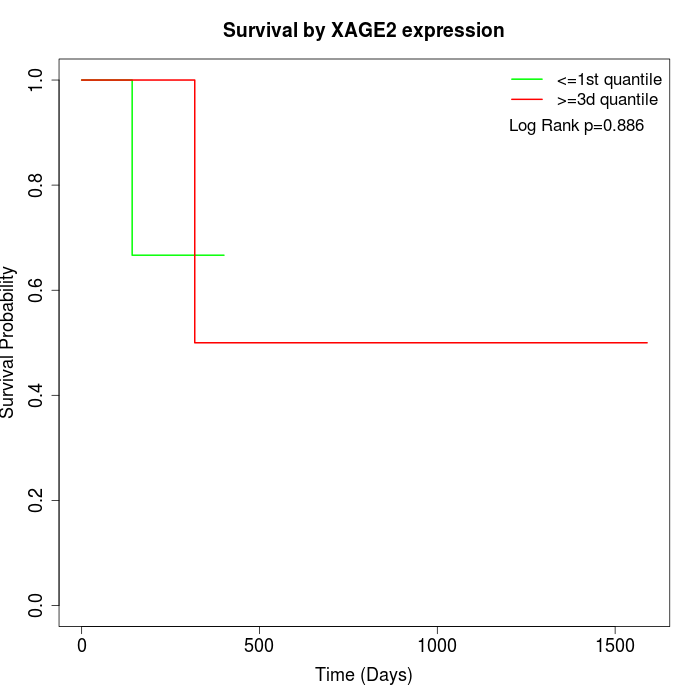
Survival by XAGE2 expression:
Note: Click image to view full size file.
Copy number change of XAGE2:
No record found for this gene.
Somatic mutations of XAGE2:
Generating mutation plots.
Highly correlated genes for XAGE2:
Showing top 20/176 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| XAGE2 | CADPS | 0.729983 | 3 | 0 | 3 |
| XAGE2 | MIRLET7BHG | 0.709241 | 3 | 0 | 3 |
| XAGE2 | TSPAN16 | 0.702393 | 3 | 0 | 3 |
| XAGE2 | PNMA3 | 0.695445 | 4 | 0 | 4 |
| XAGE2 | LINC00304 | 0.691054 | 3 | 0 | 3 |
| XAGE2 | TSSK3 | 0.680551 | 3 | 0 | 3 |
| XAGE2 | FAM30A | 0.676899 | 3 | 0 | 3 |
| XAGE2 | CLECL1 | 0.676461 | 3 | 0 | 3 |
| XAGE2 | DSCR10 | 0.674673 | 3 | 0 | 3 |
| XAGE2 | SLC22A31 | 0.669216 | 4 | 0 | 4 |
| XAGE2 | KCNQ4 | 0.665734 | 4 | 0 | 3 |
| XAGE2 | MYOD1 | 0.654961 | 4 | 0 | 3 |
| XAGE2 | DEFB128 | 0.652723 | 3 | 0 | 3 |
| XAGE2 | RBPJL | 0.651833 | 4 | 0 | 4 |
| XAGE2 | FAM71F1 | 0.650901 | 3 | 0 | 3 |
| XAGE2 | GRIK3 | 0.646983 | 3 | 0 | 3 |
| XAGE2 | SLC22A7 | 0.641245 | 4 | 0 | 3 |
| XAGE2 | TCF23 | 0.641207 | 3 | 0 | 3 |
| XAGE2 | GPR61 | 0.636881 | 4 | 0 | 3 |
| XAGE2 | TERT | 0.636 | 5 | 0 | 3 |
For details and further investigation, click here