| Full name: hedgehog acyltransferase like | Alias Symbol: KIAA1173|OACT3|MSTP002|MBOAT3 | ||
| Type: protein-coding gene | Cytoband: 3p22.1 | ||
| Entrez ID: 57467 | HGNC ID: HGNC:13242 | Ensembl Gene: ENSG00000010282 | OMIM ID: 608116 |
| Drug and gene relationship at DGIdb | |||
Expression of HHATL:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HHATL | 57467 | 223572_at | 0.0223 | 0.9484 | |
| GSE26886 | HHATL | 57467 | 223572_at | 0.2693 | 0.0185 | |
| GSE45670 | HHATL | 57467 | 223572_at | 0.1684 | 0.0446 | |
| GSE53622 | HHATL | 57467 | 88148 | -0.4605 | 0.0000 | |
| GSE53624 | HHATL | 57467 | 88148 | -0.5020 | 0.0016 | |
| GSE63941 | HHATL | 57467 | 223572_at | 0.0089 | 0.9641 | |
| GSE77861 | HHATL | 57467 | 223572_at | -0.1203 | 0.1903 | |
| GSE97050 | HHATL | 57467 | A_23_P255298 | -0.0362 | 0.8682 |
Upregulated datasets: 0; Downregulated datasets: 0.
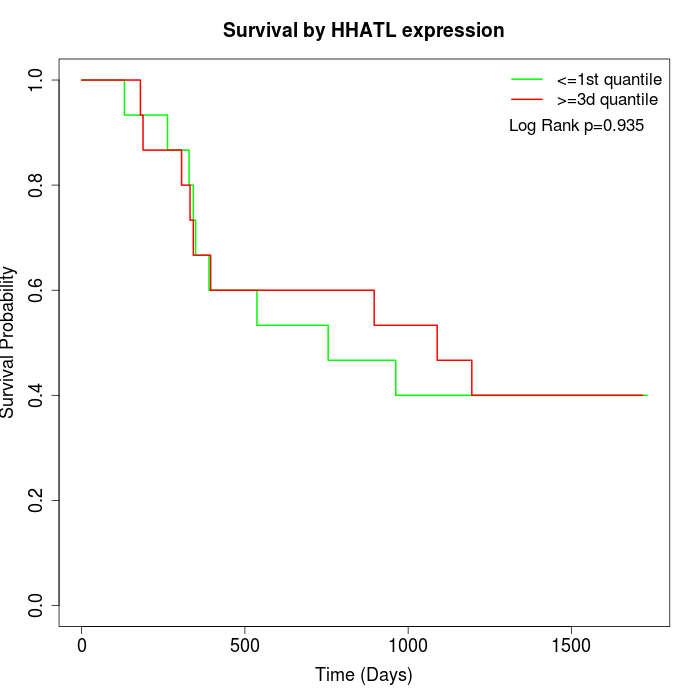
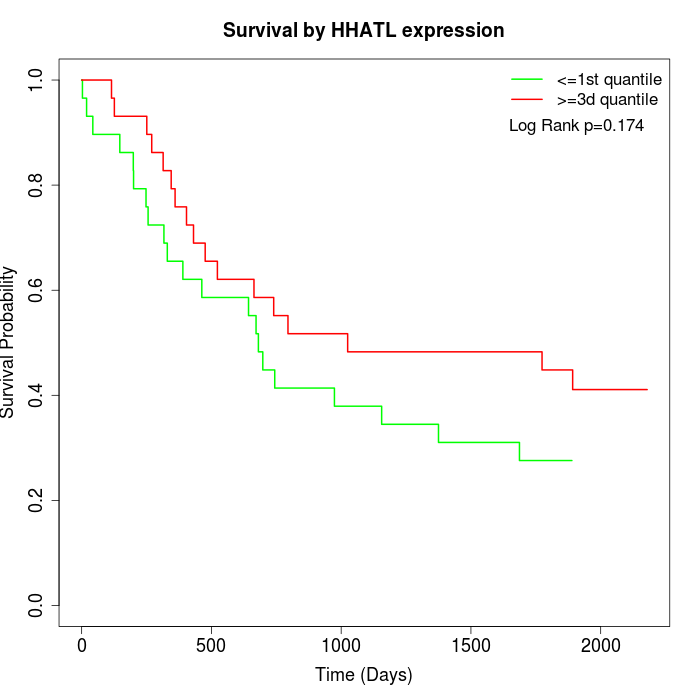
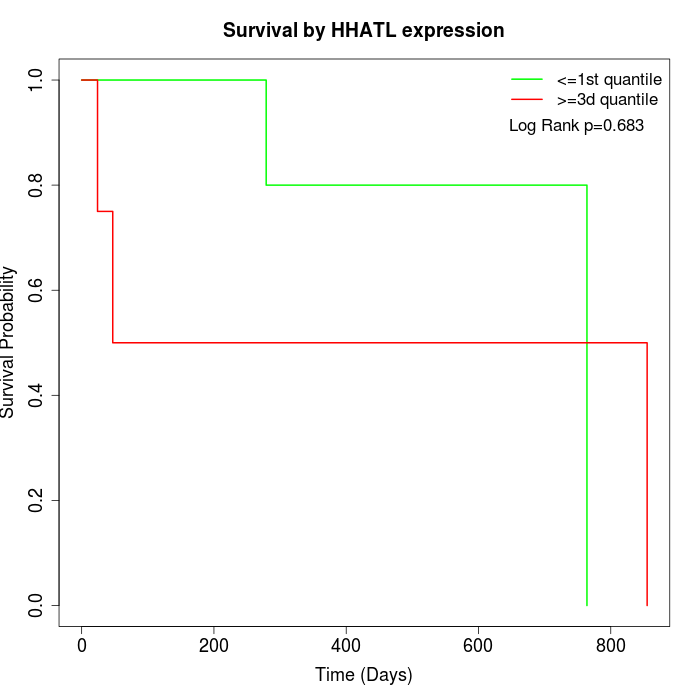
Survival by HHATL expression:
Note: Click image to view full size file.
Copy number change of HHATL:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HHATL | 57467 | 0 | 19 | 11 | |
| GSE20123 | HHATL | 57467 | 0 | 19 | 11 | |
| GSE43470 | HHATL | 57467 | 0 | 21 | 22 | |
| GSE46452 | HHATL | 57467 | 2 | 17 | 40 | |
| GSE47630 | HHATL | 57467 | 1 | 24 | 15 | |
| GSE54993 | HHATL | 57467 | 7 | 2 | 61 | |
| GSE54994 | HHATL | 57467 | 1 | 34 | 18 | |
| GSE60625 | HHATL | 57467 | 5 | 0 | 6 | |
| GSE74703 | HHATL | 57467 | 0 | 17 | 19 | |
| GSE74704 | HHATL | 57467 | 0 | 13 | 7 | |
| TCGA | HHATL | 57467 | 1 | 71 | 24 |
Total number of gains: 17; Total number of losses: 237; Total Number of normals: 234.
Somatic mutations of HHATL:
Generating mutation plots.
Highly correlated genes for HHATL:
Showing top 20/170 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HHATL | UCMA | 0.765851 | 3 | 0 | 3 |
| HHATL | ZNF235 | 0.75554 | 3 | 0 | 3 |
| HHATL | KBTBD13 | 0.73436 | 3 | 0 | 3 |
| HHATL | OR10G3 | 0.728035 | 3 | 0 | 3 |
| HHATL | KCP | 0.726459 | 3 | 0 | 3 |
| HHATL | PPP1R14D | 0.718124 | 3 | 0 | 3 |
| HHATL | ALX1 | 0.715796 | 3 | 0 | 3 |
| HHATL | PVALB | 0.712346 | 4 | 0 | 4 |
| HHATL | OR51T1 | 0.710846 | 3 | 0 | 3 |
| HHATL | VWA5B2 | 0.710307 | 4 | 0 | 3 |
| HHATL | SRRM3 | 0.709336 | 4 | 0 | 4 |
| HHATL | TSPAN11 | 0.696171 | 3 | 0 | 3 |
| HHATL | EVX1 | 0.693457 | 3 | 0 | 3 |
| HHATL | CSHL1 | 0.687544 | 4 | 0 | 4 |
| HHATL | MAPK15 | 0.687259 | 4 | 0 | 4 |
| HHATL | ASCL5 | 0.683667 | 3 | 0 | 3 |
| HHATL | CTSE | 0.678463 | 3 | 0 | 3 |
| HHATL | GPD1 | 0.674306 | 4 | 0 | 3 |
| HHATL | LRRC46 | 0.673916 | 4 | 0 | 3 |
| HHATL | GHRHR | 0.67234 | 5 | 0 | 4 |
For details and further investigation, click here