| Full name: parvalbumin | Alias Symbol: D22S749 | ||
| Type: protein-coding gene | Cytoband: 22q12.3 | ||
| Entrez ID: 5816 | HGNC ID: HGNC:9704 | Ensembl Gene: ENSG00000100362 | OMIM ID: 168890 |
| Drug and gene relationship at DGIdb | |||
Expression of PVALB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PVALB | 5816 | 205336_at | 0.0583 | 0.8752 | |
| GSE20347 | PVALB | 5816 | 205336_at | 0.0949 | 0.3272 | |
| GSE23400 | PVALB | 5816 | 205336_at | -0.0574 | 0.2571 | |
| GSE26886 | PVALB | 5816 | 205336_at | 0.2070 | 0.0494 | |
| GSE29001 | PVALB | 5816 | 205336_at | 0.0886 | 0.6418 | |
| GSE38129 | PVALB | 5816 | 205336_at | 0.0101 | 0.9410 | |
| GSE45670 | PVALB | 5816 | 205336_at | 0.2452 | 0.3871 | |
| GSE53622 | PVALB | 5816 | 111558 | 0.0967 | 0.2402 | |
| GSE53624 | PVALB | 5816 | 111558 | -0.0550 | 0.5893 | |
| GSE63941 | PVALB | 5816 | 205336_at | 0.2861 | 0.0788 | |
| GSE77861 | PVALB | 5816 | 205336_at | -0.0646 | 0.7266 | |
| GSE97050 | PVALB | 5816 | A_23_P17844 | -0.1627 | 0.6260 | |
| TCGA | PVALB | 5816 | RNAseq | -0.4107 | 0.6977 |
Upregulated datasets: 0; Downregulated datasets: 0.
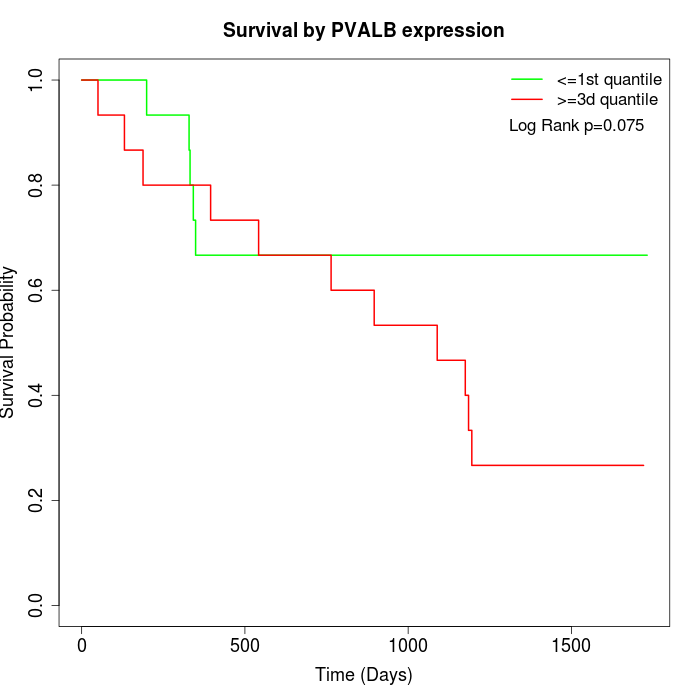
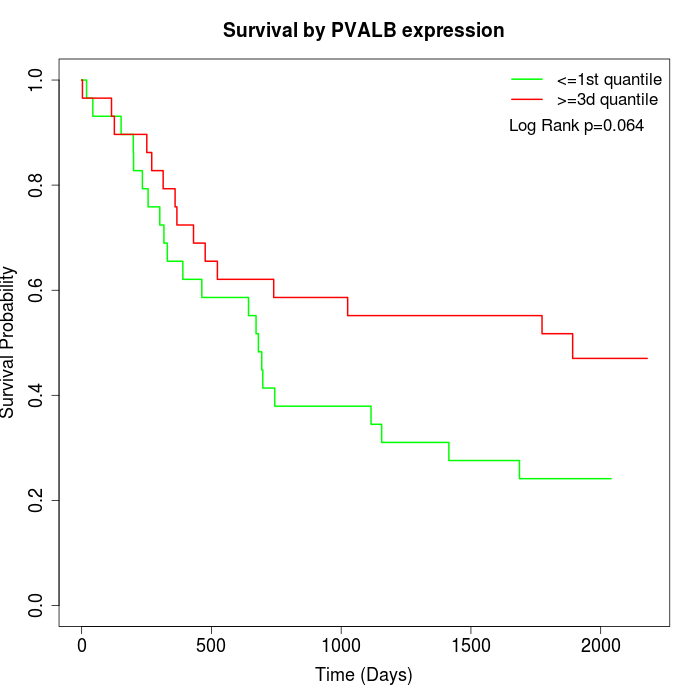
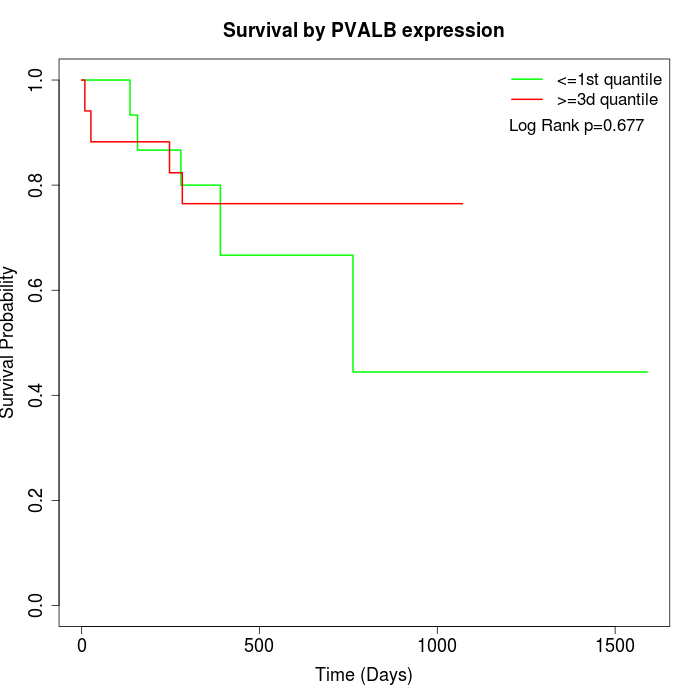
Survival by PVALB expression:
Note: Click image to view full size file.
Copy number change of PVALB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PVALB | 5816 | 6 | 6 | 18 | |
| GSE20123 | PVALB | 5816 | 6 | 5 | 19 | |
| GSE43470 | PVALB | 5816 | 4 | 6 | 33 | |
| GSE46452 | PVALB | 5816 | 31 | 1 | 27 | |
| GSE47630 | PVALB | 5816 | 8 | 5 | 27 | |
| GSE54993 | PVALB | 5816 | 3 | 6 | 61 | |
| GSE54994 | PVALB | 5816 | 10 | 9 | 34 | |
| GSE60625 | PVALB | 5816 | 5 | 0 | 6 | |
| GSE74703 | PVALB | 5816 | 4 | 4 | 28 | |
| GSE74704 | PVALB | 5816 | 3 | 2 | 15 | |
| TCGA | PVALB | 5816 | 27 | 16 | 53 |
Total number of gains: 107; Total number of losses: 60; Total Number of normals: 321.
Somatic mutations of PVALB:
Generating mutation plots.
Highly correlated genes for PVALB:
Showing top 20/413 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PVALB | CHMP6 | 0.863454 | 3 | 0 | 3 |
| PVALB | SPACA3 | 0.822954 | 3 | 0 | 3 |
| PVALB | DCDC1 | 0.798914 | 3 | 0 | 3 |
| PVALB | LILRA3 | 0.787379 | 3 | 0 | 3 |
| PVALB | IL1F10 | 0.775252 | 3 | 0 | 3 |
| PVALB | OSM | 0.770478 | 3 | 0 | 3 |
| PVALB | STRC | 0.769862 | 3 | 0 | 3 |
| PVALB | MAG | 0.762104 | 5 | 0 | 5 |
| PVALB | TUSC1 | 0.756964 | 3 | 0 | 3 |
| PVALB | NCAN | 0.746505 | 3 | 0 | 3 |
| PVALB | PABPN1L | 0.744101 | 3 | 0 | 3 |
| PVALB | C19orf25 | 0.742206 | 3 | 0 | 3 |
| PVALB | SLC6A18 | 0.729616 | 3 | 0 | 3 |
| PVALB | TMEM179 | 0.724698 | 3 | 0 | 3 |
| PVALB | FBXL20 | 0.724513 | 3 | 0 | 3 |
| PVALB | SLC39A5 | 0.723689 | 4 | 0 | 3 |
| PVALB | MOG | 0.719964 | 4 | 0 | 4 |
| PVALB | KCNH3 | 0.719315 | 4 | 0 | 4 |
| PVALB | ZNF843 | 0.715735 | 4 | 0 | 4 |
| PVALB | SLC22A31 | 0.713819 | 3 | 0 | 3 |
For details and further investigation, click here