| Full name: insulin degrading enzyme | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q23.33 | ||
| Entrez ID: 3416 | HGNC ID: HGNC:5381 | Ensembl Gene: ENSG00000119912 | OMIM ID: 146680 |
| Drug and gene relationship at DGIdb | |||
Expression of IDE:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | IDE | 3416 | 203327_at | -0.4432 | 0.1464 | |
| GSE20347 | IDE | 3416 | 203327_at | -0.7320 | 0.0000 | |
| GSE23400 | IDE | 3416 | 217496_s_at | -0.1499 | 0.0108 | |
| GSE26886 | IDE | 3416 | 203327_at | -0.6096 | 0.0126 | |
| GSE29001 | IDE | 3416 | 203327_at | -0.8102 | 0.0193 | |
| GSE38129 | IDE | 3416 | 203327_at | -0.5671 | 0.0000 | |
| GSE45670 | IDE | 3416 | 203327_at | -0.4558 | 0.0004 | |
| GSE53622 | IDE | 3416 | 56810 | -0.4777 | 0.0000 | |
| GSE53624 | IDE | 3416 | 56810 | -0.4623 | 0.0000 | |
| GSE63941 | IDE | 3416 | 217496_s_at | 0.6949 | 0.0459 | |
| GSE77861 | IDE | 3416 | 203327_at | -0.5062 | 0.1365 | |
| GSE97050 | IDE | 3416 | A_23_P63681 | -0.0823 | 0.7358 | |
| SRP007169 | IDE | 3416 | RNAseq | -1.2042 | 0.0033 | |
| SRP008496 | IDE | 3416 | RNAseq | -0.7514 | 0.0093 | |
| SRP064894 | IDE | 3416 | RNAseq | -0.5689 | 0.0001 | |
| SRP133303 | IDE | 3416 | RNAseq | -0.1410 | 0.4373 | |
| SRP159526 | IDE | 3416 | RNAseq | -0.3651 | 0.3503 | |
| SRP193095 | IDE | 3416 | RNAseq | -0.3624 | 0.0001 | |
| SRP219564 | IDE | 3416 | RNAseq | -0.3888 | 0.0710 | |
| TCGA | IDE | 3416 | RNAseq | -0.0883 | 0.1146 |
Upregulated datasets: 0; Downregulated datasets: 1.
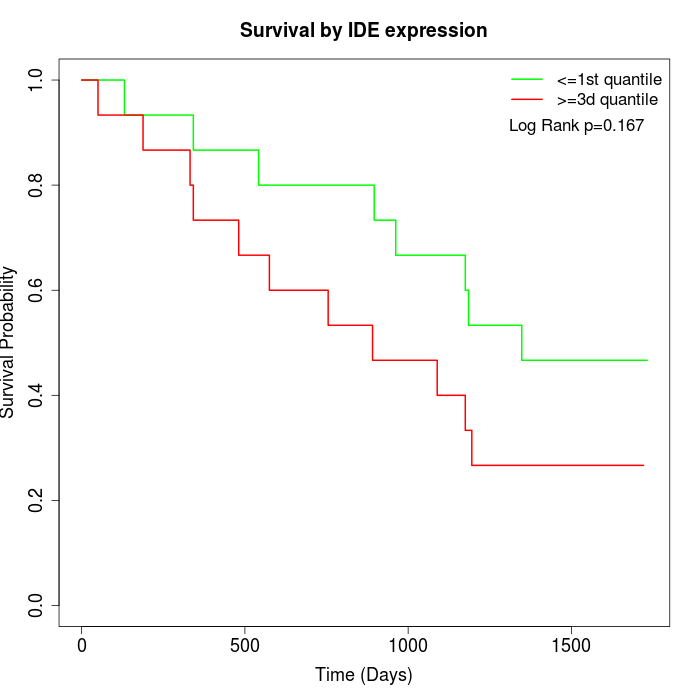
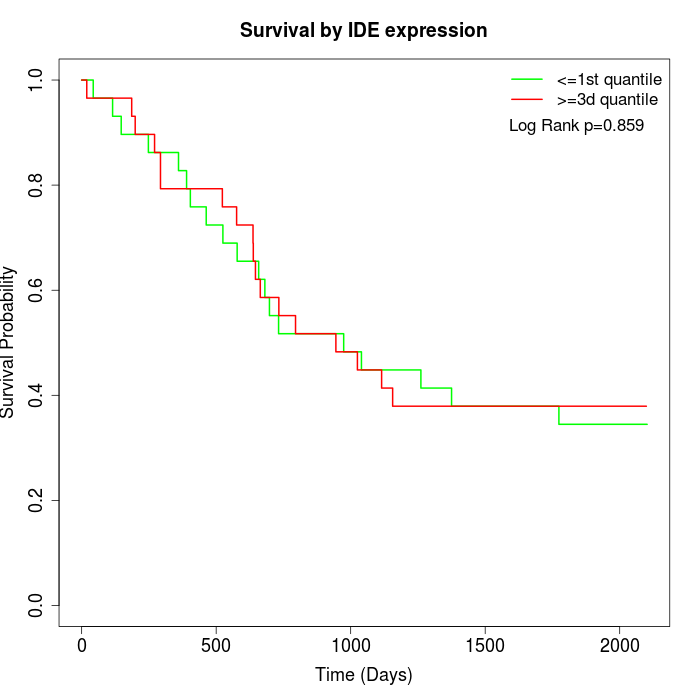
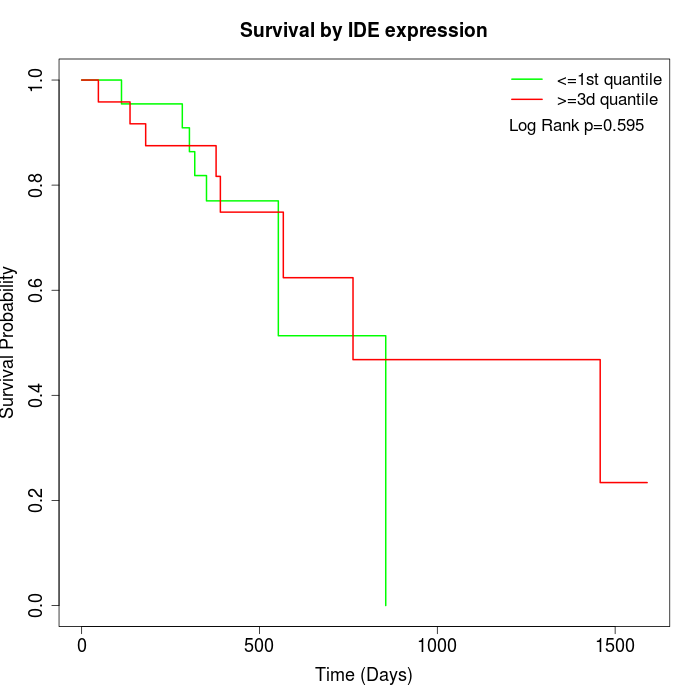
Survival by IDE expression:
Note: Click image to view full size file.
Copy number change of IDE:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | IDE | 3416 | 2 | 7 | 21 | |
| GSE20123 | IDE | 3416 | 2 | 6 | 22 | |
| GSE43470 | IDE | 3416 | 0 | 8 | 35 | |
| GSE46452 | IDE | 3416 | 0 | 11 | 48 | |
| GSE47630 | IDE | 3416 | 2 | 14 | 24 | |
| GSE54993 | IDE | 3416 | 7 | 0 | 63 | |
| GSE54994 | IDE | 3416 | 1 | 10 | 42 | |
| GSE60625 | IDE | 3416 | 0 | 0 | 11 | |
| GSE74703 | IDE | 3416 | 0 | 6 | 30 | |
| GSE74704 | IDE | 3416 | 0 | 4 | 16 | |
| TCGA | IDE | 3416 | 7 | 26 | 63 |
Total number of gains: 21; Total number of losses: 92; Total Number of normals: 375.
Somatic mutations of IDE:
Generating mutation plots.
Highly correlated genes for IDE:
Showing top 20/1119 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| IDE | CGRRF1 | 0.786287 | 3 | 0 | 3 |
| IDE | ARSG | 0.759888 | 3 | 0 | 3 |
| IDE | GDAP2 | 0.746477 | 3 | 0 | 3 |
| IDE | TMEM126A | 0.737514 | 4 | 0 | 4 |
| IDE | VDAC2 | 0.729014 | 9 | 0 | 8 |
| IDE | SARNP | 0.717535 | 3 | 0 | 3 |
| IDE | MTMR10 | 0.713056 | 6 | 0 | 6 |
| IDE | ZNF480 | 0.703527 | 4 | 0 | 3 |
| IDE | CCIN | 0.701775 | 4 | 0 | 3 |
| IDE | AP3M1 | 0.698363 | 6 | 0 | 5 |
| IDE | PSMD8 | 0.695776 | 4 | 0 | 3 |
| IDE | OSBPL2 | 0.695439 | 4 | 0 | 4 |
| IDE | ZNF585A | 0.68933 | 4 | 0 | 3 |
| IDE | BLOC1S1 | 0.688481 | 8 | 0 | 7 |
| IDE | ZNF529 | 0.683859 | 3 | 0 | 3 |
| IDE | RBM20 | 0.683195 | 4 | 0 | 3 |
| IDE | MPC1 | 0.683056 | 9 | 0 | 8 |
| IDE | ALG13 | 0.682572 | 4 | 0 | 3 |
| IDE | C12orf29 | 0.681568 | 10 | 0 | 9 |
| IDE | GAPVD1 | 0.681047 | 8 | 0 | 8 |
For details and further investigation, click here