| Full name: ganglioside induced differentiation associated protein 2 | Alias Symbol: FLJ20142|dJ776P7.1|MACROD3 | ||
| Type: protein-coding gene | Cytoband: 1p12 | ||
| Entrez ID: 54834 | HGNC ID: HGNC:18010 | Ensembl Gene: ENSG00000196505 | OMIM ID: 618128 |
| Drug and gene relationship at DGIdb | |||
Expression of GDAP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GDAP2 | 54834 | 1554154_at | -0.1336 | 0.8094 | |
| GSE20347 | GDAP2 | 54834 | 219473_at | 0.0518 | 0.6222 | |
| GSE23400 | GDAP2 | 54834 | 219473_at | -0.1519 | 0.0002 | |
| GSE26886 | GDAP2 | 54834 | 219473_at | -0.6857 | 0.0050 | |
| GSE29001 | GDAP2 | 54834 | 219473_at | -0.1026 | 0.7861 | |
| GSE38129 | GDAP2 | 54834 | 219473_at | -0.0453 | 0.6355 | |
| GSE45670 | GDAP2 | 54834 | 1554154_at | -0.1015 | 0.5891 | |
| GSE53622 | GDAP2 | 54834 | 165572 | -0.1382 | 0.0679 | |
| GSE53624 | GDAP2 | 54834 | 165572 | 0.1178 | 0.1155 | |
| GSE63941 | GDAP2 | 54834 | 1554154_at | 1.2649 | 0.0544 | |
| GSE77861 | GDAP2 | 54834 | 1554154_at | 0.3485 | 0.0835 | |
| GSE97050 | GDAP2 | 54834 | A_24_P233878 | -0.2193 | 0.3725 | |
| SRP007169 | GDAP2 | 54834 | RNAseq | -0.2605 | 0.5143 | |
| SRP008496 | GDAP2 | 54834 | RNAseq | -0.4351 | 0.0426 | |
| SRP064894 | GDAP2 | 54834 | RNAseq | 0.0123 | 0.9533 | |
| SRP133303 | GDAP2 | 54834 | RNAseq | 0.0585 | 0.5366 | |
| SRP159526 | GDAP2 | 54834 | RNAseq | -0.4596 | 0.0691 | |
| SRP193095 | GDAP2 | 54834 | RNAseq | -0.1662 | 0.0167 | |
| SRP219564 | GDAP2 | 54834 | RNAseq | 0.0082 | 0.9886 | |
| TCGA | GDAP2 | 54834 | RNAseq | -0.1711 | 0.0017 |
Upregulated datasets: 0; Downregulated datasets: 0.
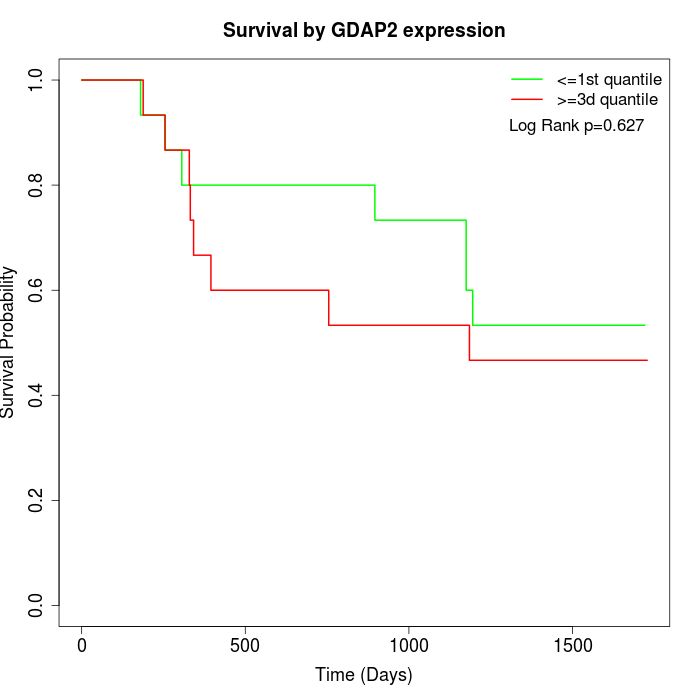
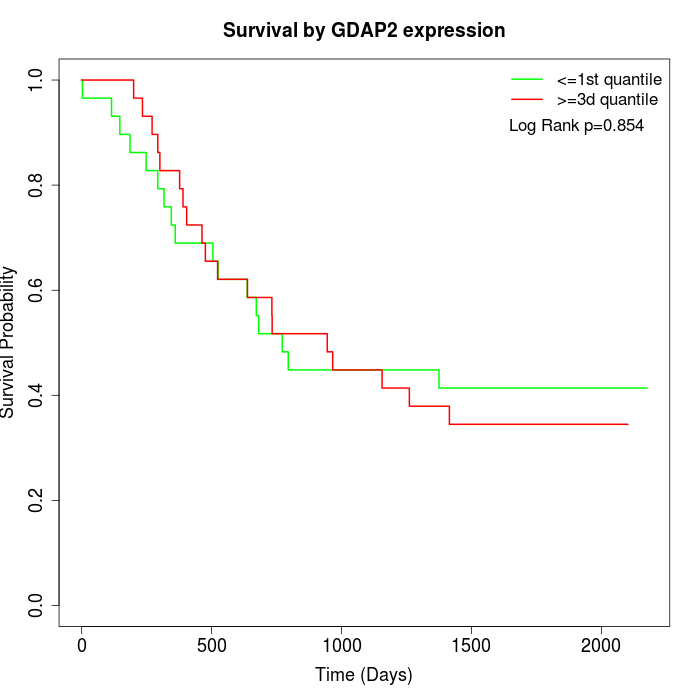
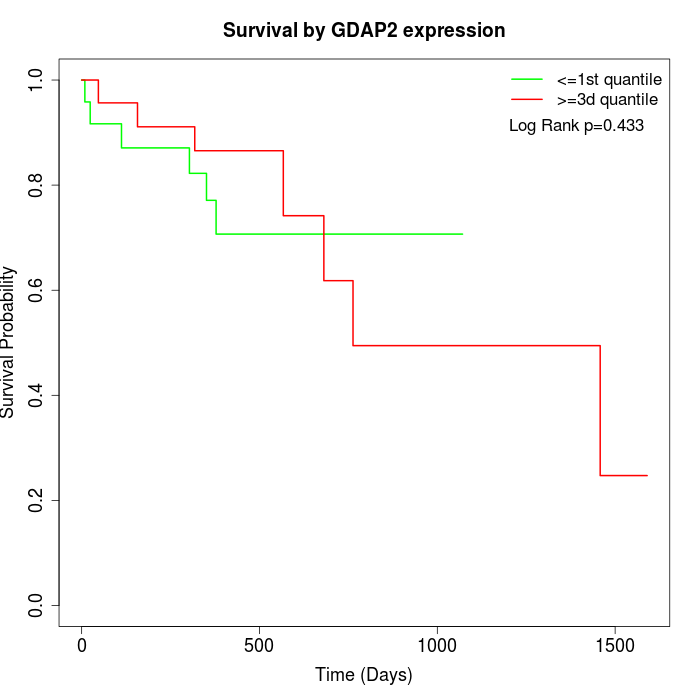
Survival by GDAP2 expression:
Note: Click image to view full size file.
Copy number change of GDAP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GDAP2 | 54834 | 0 | 8 | 22 | |
| GSE20123 | GDAP2 | 54834 | 0 | 8 | 22 | |
| GSE43470 | GDAP2 | 54834 | 0 | 3 | 40 | |
| GSE46452 | GDAP2 | 54834 | 2 | 1 | 56 | |
| GSE47630 | GDAP2 | 54834 | 9 | 5 | 26 | |
| GSE54993 | GDAP2 | 54834 | 0 | 1 | 69 | |
| GSE54994 | GDAP2 | 54834 | 11 | 1 | 41 | |
| GSE60625 | GDAP2 | 54834 | 0 | 0 | 11 | |
| GSE74703 | GDAP2 | 54834 | 0 | 3 | 33 | |
| GSE74704 | GDAP2 | 54834 | 0 | 4 | 16 | |
| TCGA | GDAP2 | 54834 | 15 | 28 | 53 |
Total number of gains: 37; Total number of losses: 62; Total Number of normals: 389.
Somatic mutations of GDAP2:
Generating mutation plots.
Highly correlated genes for GDAP2:
Showing top 20/199 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GDAP2 | PCNT | 0.777302 | 3 | 0 | 3 |
| GDAP2 | AKAP17A | 0.765022 | 3 | 0 | 3 |
| GDAP2 | IMPACT | 0.764169 | 3 | 0 | 3 |
| GDAP2 | NIPSNAP3A | 0.758506 | 3 | 0 | 3 |
| GDAP2 | ZFP3 | 0.755157 | 3 | 0 | 3 |
| GDAP2 | MOB4 | 0.750354 | 3 | 0 | 3 |
| GDAP2 | GGA3 | 0.747724 | 3 | 0 | 3 |
| GDAP2 | IDE | 0.746477 | 3 | 0 | 3 |
| GDAP2 | DNA2 | 0.731815 | 3 | 0 | 3 |
| GDAP2 | PANK1 | 0.731399 | 3 | 0 | 3 |
| GDAP2 | MYSM1 | 0.726542 | 3 | 0 | 3 |
| GDAP2 | HDDC2 | 0.725212 | 3 | 0 | 3 |
| GDAP2 | BTAF1 | 0.724509 | 4 | 0 | 3 |
| GDAP2 | TRAPPC8 | 0.721614 | 4 | 0 | 3 |
| GDAP2 | SASS6 | 0.720646 | 3 | 0 | 3 |
| GDAP2 | TRAPPC2 | 0.719189 | 4 | 0 | 4 |
| GDAP2 | SLC30A7 | 0.718609 | 3 | 0 | 3 |
| GDAP2 | ACAP2 | 0.717205 | 3 | 0 | 3 |
| GDAP2 | ANKIB1 | 0.71709 | 3 | 0 | 3 |
| GDAP2 | USP24 | 0.716266 | 3 | 0 | 3 |
For details and further investigation, click here