| Full name: voltage dependent anion channel 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q22.2 | ||
| Entrez ID: 7417 | HGNC ID: HGNC:12672 | Ensembl Gene: ENSG00000165637 | OMIM ID: 193245 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
VDAC2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04022 | cGMP-PKG signaling pathway | |
| hsa05012 | Parkinson's disease |
Expression of VDAC2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | VDAC2 | 7417 | 211662_s_at | -0.3687 | 0.2143 | |
| GSE20347 | VDAC2 | 7417 | 211662_s_at | -0.6225 | 0.0000 | |
| GSE23400 | VDAC2 | 7417 | 211662_s_at | -0.4655 | 0.0000 | |
| GSE26886 | VDAC2 | 7417 | 211662_s_at | -0.7381 | 0.0001 | |
| GSE29001 | VDAC2 | 7417 | 211662_s_at | -0.8856 | 0.0044 | |
| GSE38129 | VDAC2 | 7417 | 211662_s_at | -0.3661 | 0.0126 | |
| GSE45670 | VDAC2 | 7417 | 211662_s_at | -0.3316 | 0.0078 | |
| GSE53622 | VDAC2 | 7417 | 148246 | -0.6675 | 0.0000 | |
| GSE53624 | VDAC2 | 7417 | 34852 | -0.4394 | 0.0000 | |
| GSE63941 | VDAC2 | 7417 | 211662_s_at | 0.4098 | 0.0669 | |
| GSE77861 | VDAC2 | 7417 | 211662_s_at | -0.2400 | 0.2702 | |
| GSE97050 | VDAC2 | 7417 | A_33_P3415037 | -0.2166 | 0.4067 | |
| SRP007169 | VDAC2 | 7417 | RNAseq | -1.4425 | 0.0017 | |
| SRP008496 | VDAC2 | 7417 | RNAseq | -1.2433 | 0.0000 | |
| SRP064894 | VDAC2 | 7417 | RNAseq | -1.2109 | 0.0000 | |
| SRP133303 | VDAC2 | 7417 | RNAseq | -0.3363 | 0.0680 | |
| SRP159526 | VDAC2 | 7417 | RNAseq | -0.7282 | 0.1165 | |
| SRP193095 | VDAC2 | 7417 | RNAseq | -0.7318 | 0.0000 | |
| SRP219564 | VDAC2 | 7417 | RNAseq | -0.9407 | 0.0017 | |
| TCGA | VDAC2 | 7417 | RNAseq | 0.0556 | 0.2976 |
Upregulated datasets: 0; Downregulated datasets: 3.
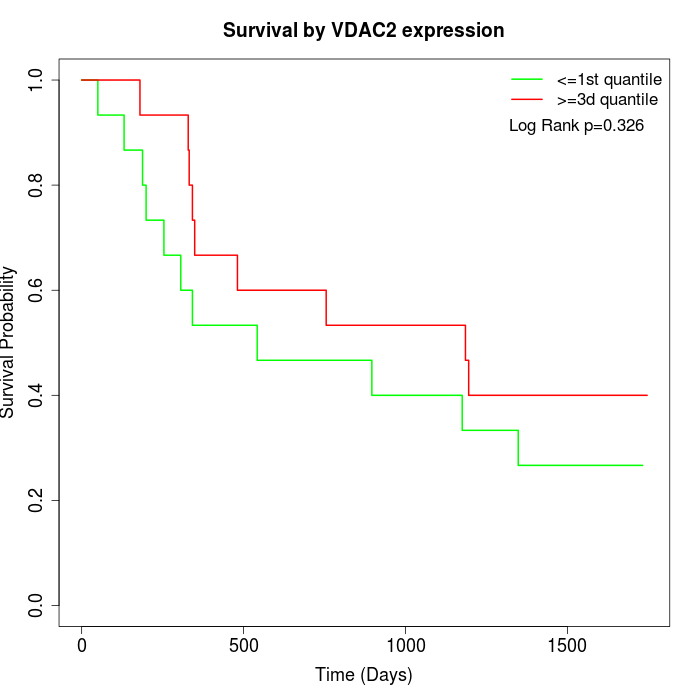
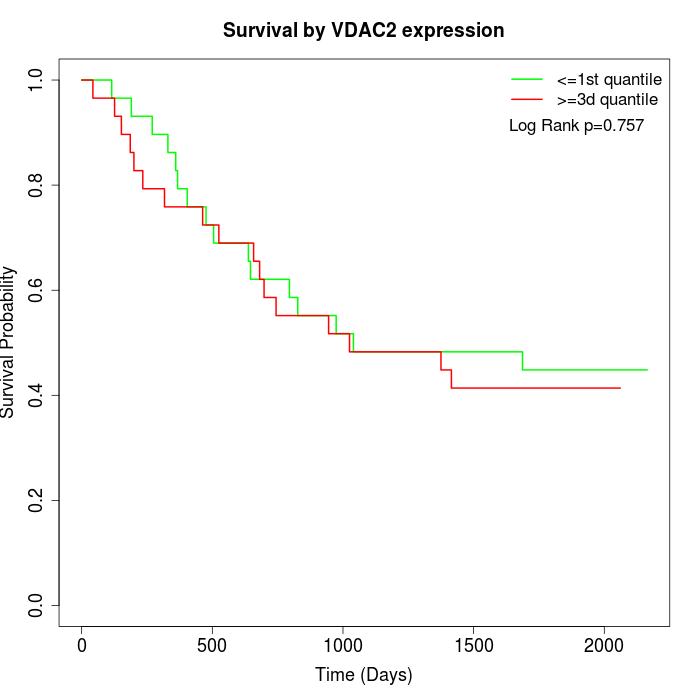
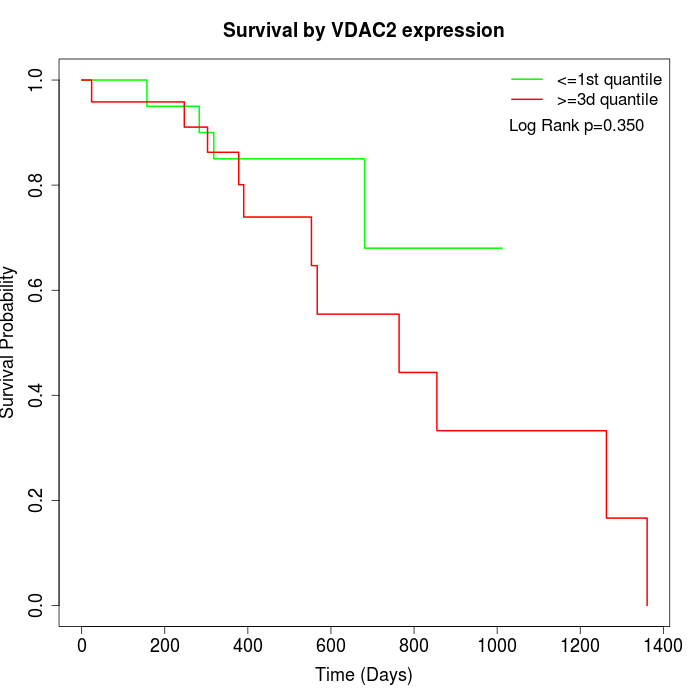
Survival by VDAC2 expression:
Note: Click image to view full size file.
Copy number change of VDAC2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VDAC2 | 7417 | 3 | 5 | 22 | |
| GSE20123 | VDAC2 | 7417 | 3 | 4 | 23 | |
| GSE43470 | VDAC2 | 7417 | 2 | 7 | 34 | |
| GSE46452 | VDAC2 | 7417 | 0 | 11 | 48 | |
| GSE47630 | VDAC2 | 7417 | 2 | 14 | 24 | |
| GSE54993 | VDAC2 | 7417 | 7 | 0 | 63 | |
| GSE54994 | VDAC2 | 7417 | 2 | 10 | 41 | |
| GSE60625 | VDAC2 | 7417 | 0 | 0 | 11 | |
| GSE74703 | VDAC2 | 7417 | 2 | 4 | 30 | |
| GSE74704 | VDAC2 | 7417 | 1 | 2 | 17 | |
| TCGA | VDAC2 | 7417 | 11 | 23 | 62 |
Total number of gains: 33; Total number of losses: 80; Total Number of normals: 375.
Somatic mutations of VDAC2:
Generating mutation plots.
Highly correlated genes for VDAC2:
Showing top 20/1480 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VDAC2 | ZFC3H1 | 0.802715 | 3 | 0 | 3 |
| VDAC2 | PUM2 | 0.762562 | 3 | 0 | 3 |
| VDAC2 | ZNF740 | 0.758468 | 3 | 0 | 3 |
| VDAC2 | ARIH1 | 0.752764 | 4 | 0 | 4 |
| VDAC2 | CCNDBP1 | 0.751928 | 5 | 0 | 5 |
| VDAC2 | MFAP1 | 0.748084 | 3 | 0 | 3 |
| VDAC2 | CCNYL1 | 0.738764 | 4 | 0 | 4 |
| VDAC2 | DCTD | 0.73288 | 3 | 0 | 3 |
| VDAC2 | SAMD8 | 0.730066 | 7 | 0 | 7 |
| VDAC2 | IDE | 0.729014 | 9 | 0 | 8 |
| VDAC2 | BLOC1S2 | 0.72722 | 4 | 0 | 3 |
| VDAC2 | SMAGP | 0.720532 | 10 | 0 | 10 |
| VDAC2 | MFSD5 | 0.719311 | 10 | 0 | 9 |
| VDAC2 | CIAPIN1 | 0.714491 | 3 | 0 | 3 |
| VDAC2 | DDI2 | 0.713885 | 3 | 0 | 3 |
| VDAC2 | TMEM218 | 0.713844 | 3 | 0 | 3 |
| VDAC2 | FBXO8 | 0.713269 | 4 | 0 | 4 |
| VDAC2 | HEATR5B | 0.709856 | 3 | 0 | 3 |
| VDAC2 | SOCS6 | 0.708747 | 8 | 0 | 8 |
| VDAC2 | PRPF38A | 0.708215 | 4 | 0 | 3 |
For details and further investigation, click here