| Full name: calicin | Alias Symbol: KBTBD14|BTBD20 | ||
| Type: protein-coding gene | Cytoband: 9p13.3 | ||
| Entrez ID: 881 | HGNC ID: HGNC:1568 | Ensembl Gene: ENSG00000185972 | OMIM ID: 603960 |
| Drug and gene relationship at DGIdb | |||
Expression of CCIN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCIN | 881 | 210642_at | -0.2072 | 0.2324 | |
| GSE20347 | CCIN | 881 | 210642_at | -0.3086 | 0.0000 | |
| GSE23400 | CCIN | 881 | 210642_at | -0.2588 | 0.0000 | |
| GSE26886 | CCIN | 881 | 210642_at | -0.0173 | 0.9043 | |
| GSE29001 | CCIN | 881 | 210642_at | -0.2024 | 0.1059 | |
| GSE38129 | CCIN | 881 | 210642_at | -0.3132 | 0.0001 | |
| GSE45670 | CCIN | 881 | 210642_at | -0.1132 | 0.2023 | |
| GSE53622 | CCIN | 881 | 76278 | 0.2911 | 0.0929 | |
| GSE53624 | CCIN | 881 | 76278 | -0.1184 | 0.4959 | |
| GSE63941 | CCIN | 881 | 210642_at | -0.1114 | 0.6106 | |
| GSE77861 | CCIN | 881 | 210642_at | -0.2749 | 0.0483 | |
| GSE97050 | CCIN | 881 | A_23_P60227 | -0.3674 | 0.1581 | |
| TCGA | CCIN | 881 | RNAseq | -0.9078 | 0.0435 |
Upregulated datasets: 0; Downregulated datasets: 0.
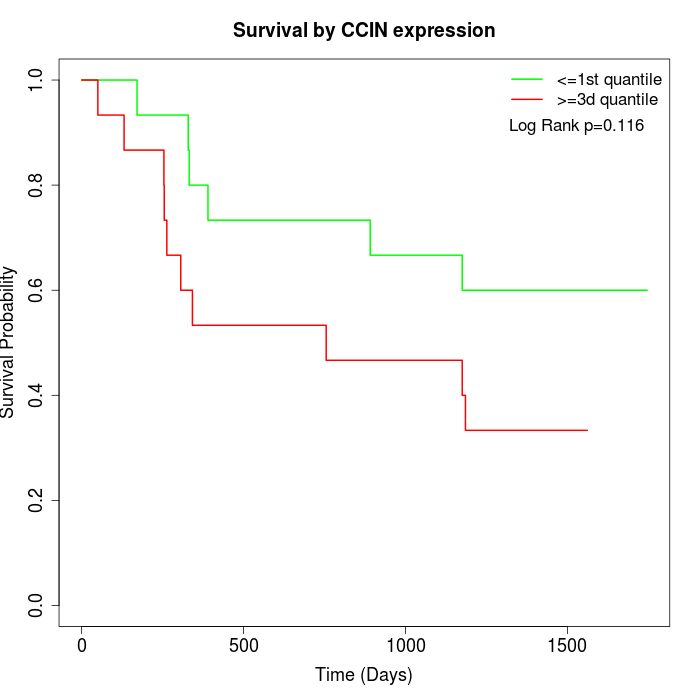
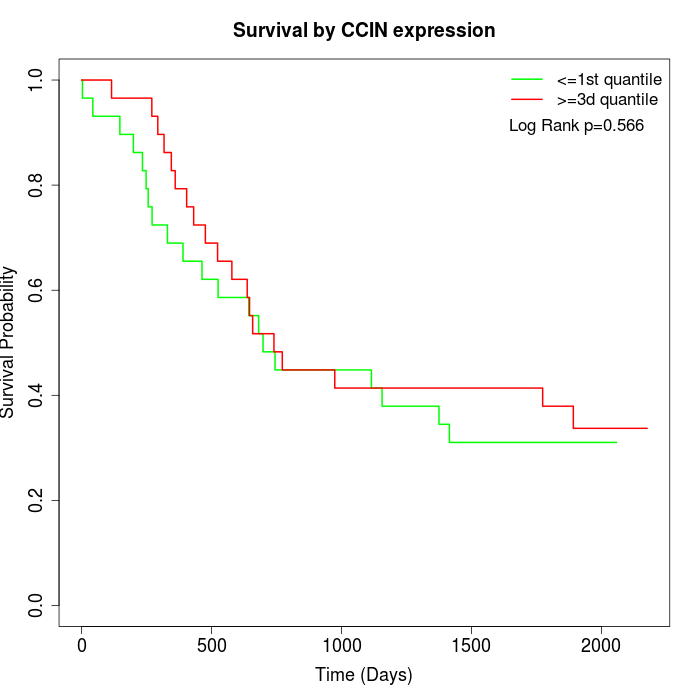
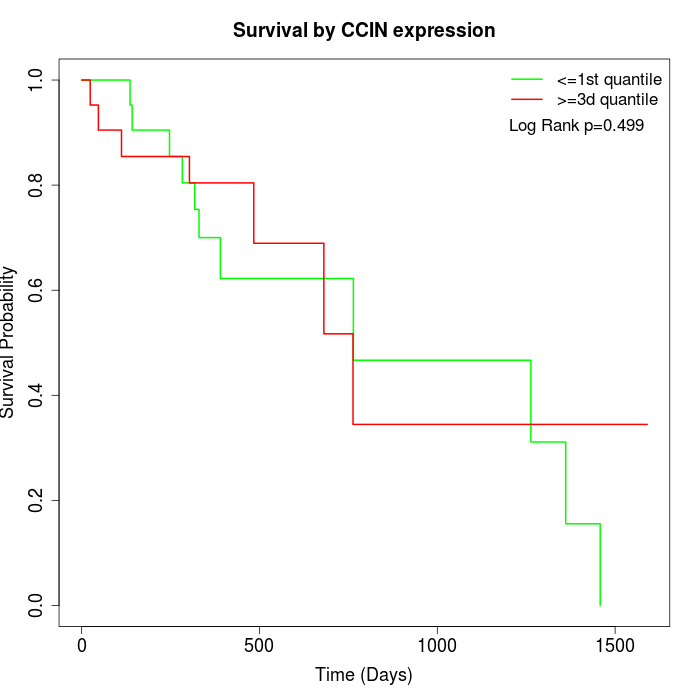
Survival by CCIN expression:
Note: Click image to view full size file.
Copy number change of CCIN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCIN | 881 | 3 | 11 | 16 | |
| GSE20123 | CCIN | 881 | 3 | 11 | 16 | |
| GSE43470 | CCIN | 881 | 3 | 10 | 30 | |
| GSE46452 | CCIN | 881 | 6 | 15 | 38 | |
| GSE47630 | CCIN | 881 | 1 | 20 | 19 | |
| GSE54993 | CCIN | 881 | 6 | 0 | 64 | |
| GSE54994 | CCIN | 881 | 5 | 12 | 36 | |
| GSE60625 | CCIN | 881 | 0 | 0 | 11 | |
| GSE74703 | CCIN | 881 | 2 | 7 | 27 | |
| GSE74704 | CCIN | 881 | 0 | 9 | 11 | |
| TCGA | CCIN | 881 | 15 | 44 | 37 |
Total number of gains: 44; Total number of losses: 139; Total Number of normals: 305.
Somatic mutations of CCIN:
Generating mutation plots.
Highly correlated genes for CCIN:
Showing top 20/1265 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCIN | SPEG | 0.901857 | 3 | 0 | 3 |
| CCIN | OR2T33 | 0.889462 | 3 | 0 | 3 |
| CCIN | OR2AT4 | 0.851888 | 3 | 0 | 3 |
| CCIN | KIR3DL2 | 0.837435 | 3 | 0 | 3 |
| CCIN | SZT2 | 0.835563 | 4 | 0 | 4 |
| CCIN | TMEM38A | 0.82503 | 3 | 0 | 3 |
| CCIN | KRTAP10-9 | 0.812821 | 3 | 0 | 3 |
| CCIN | KRTAP12-2 | 0.806326 | 3 | 0 | 3 |
| CCIN | ALG11 | 0.802917 | 3 | 0 | 3 |
| CCIN | SPRY1 | 0.802271 | 3 | 0 | 3 |
| CCIN | KRTAP10-4 | 0.799132 | 3 | 0 | 3 |
| CCIN | SVIP | 0.796573 | 3 | 0 | 3 |
| CCIN | OR1L6 | 0.794854 | 3 | 0 | 3 |
| CCIN | OTUD7B | 0.794294 | 3 | 0 | 3 |
| CCIN | SLC26A5 | 0.793253 | 3 | 0 | 3 |
| CCIN | LINC00930 | 0.789568 | 3 | 0 | 3 |
| CCIN | EIF4E3 | 0.779412 | 3 | 0 | 3 |
| CCIN | BEX4 | 0.777263 | 4 | 0 | 4 |
| CCIN | C12orf50 | 0.769657 | 3 | 0 | 3 |
| CCIN | ART1 | 0.767476 | 3 | 0 | 3 |
For details and further investigation, click here