| Full name: kelch like family member 33 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 14q11.2 | ||
| Entrez ID: 123103 | HGNC ID: HGNC:31952 | Ensembl Gene: ENSG00000185271 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of KLHL33:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | KLHL33 | 123103 | 25008 | 0.2833 | 0.0008 | |
| GSE53624 | KLHL33 | 123103 | 25008 | 0.3285 | 0.0000 | |
| GSE97050 | KLHL33 | 123103 | A_33_P3399618 | -0.0425 | 0.8543 | |
| SRP007169 | KLHL33 | 123103 | RNAseq | 0.2396 | 0.7444 | |
| SRP064894 | KLHL33 | 123103 | RNAseq | -0.5782 | 0.0328 | |
| SRP133303 | KLHL33 | 123103 | RNAseq | 0.0724 | 0.6865 | |
| SRP159526 | KLHL33 | 123103 | RNAseq | -0.8359 | 0.1179 | |
| SRP193095 | KLHL33 | 123103 | RNAseq | 0.2052 | 0.1816 | |
| SRP219564 | KLHL33 | 123103 | RNAseq | -0.1970 | 0.7371 | |
| TCGA | KLHL33 | 123103 | RNAseq | -1.8688 | 0.0131 |
Upregulated datasets: 0; Downregulated datasets: 1.
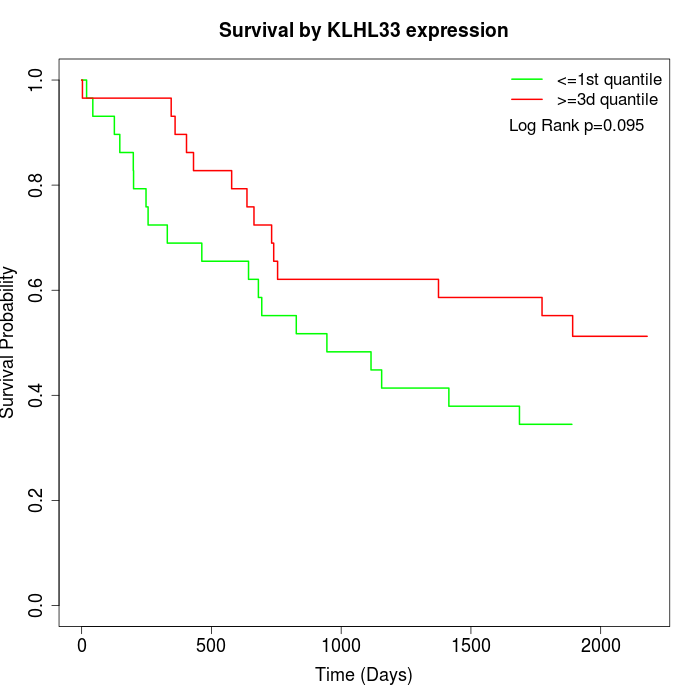
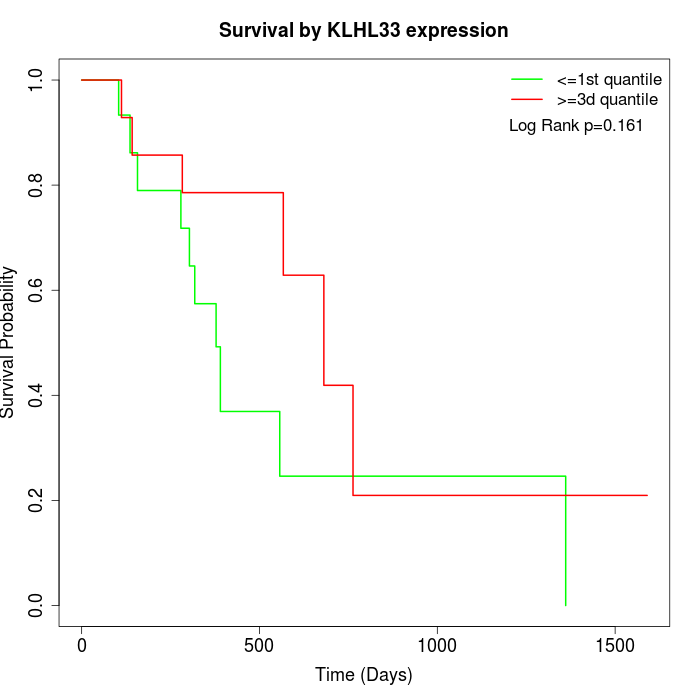
Survival by KLHL33 expression:
Note: Click image to view full size file.
Copy number change of KLHL33:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KLHL33 | 123103 | 10 | 3 | 17 | |
| GSE20123 | KLHL33 | 123103 | 8 | 3 | 19 | |
| GSE43470 | KLHL33 | 123103 | 7 | 1 | 35 | |
| GSE46452 | KLHL33 | 123103 | 19 | 2 | 38 | |
| GSE47630 | KLHL33 | 123103 | 10 | 10 | 20 | |
| GSE54993 | KLHL33 | 123103 | 3 | 11 | 56 | |
| GSE54994 | KLHL33 | 123103 | 18 | 5 | 30 | |
| GSE60625 | KLHL33 | 123103 | 0 | 2 | 9 | |
| GSE74703 | KLHL33 | 123103 | 6 | 1 | 29 | |
| GSE74704 | KLHL33 | 123103 | 4 | 2 | 14 | |
| TCGA | KLHL33 | 123103 | 28 | 16 | 52 |
Total number of gains: 113; Total number of losses: 56; Total Number of normals: 319.
Somatic mutations of KLHL33:
Generating mutation plots.
Highly correlated genes for KLHL33:
Showing top 20/55 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KLHL33 | OR2T33 | 0.764185 | 3 | 0 | 3 |
| KLHL33 | BARHL1 | 0.762521 | 3 | 0 | 3 |
| KLHL33 | CDH24 | 0.748485 | 3 | 0 | 3 |
| KLHL33 | CCIN | 0.747657 | 3 | 0 | 3 |
| KLHL33 | SPEG | 0.746405 | 3 | 0 | 3 |
| KLHL33 | TG | 0.736861 | 3 | 0 | 3 |
| KLHL33 | ALG11 | 0.736007 | 3 | 0 | 3 |
| KLHL33 | CCDC154 | 0.735789 | 3 | 0 | 3 |
| KLHL33 | TMEM63B | 0.734972 | 3 | 0 | 3 |
| KLHL33 | GRIA4 | 0.733956 | 3 | 0 | 3 |
| KLHL33 | KRTAP12-2 | 0.730488 | 3 | 0 | 3 |
| KLHL33 | PARP6 | 0.729006 | 3 | 0 | 3 |
| KLHL33 | NRL | 0.724858 | 3 | 0 | 3 |
| KLHL33 | RBM17 | 0.721267 | 3 | 0 | 3 |
| KLHL33 | SZT2 | 0.721168 | 3 | 0 | 3 |
| KLHL33 | GRIP2 | 0.71415 | 3 | 0 | 3 |
| KLHL33 | COL11A2 | 0.711213 | 3 | 0 | 3 |
| KLHL33 | ZNF248 | 0.711162 | 3 | 0 | 3 |
| KLHL33 | KRTAP10-9 | 0.70986 | 3 | 0 | 3 |
| KLHL33 | OR1D5 | 0.703961 | 3 | 0 | 3 |
For details and further investigation, click here