| Full name: keratin 79 | Alias Symbol: K6L|KRT6L | ||
| Type: protein-coding gene | Cytoband: 12q13.13 | ||
| Entrez ID: 338785 | HGNC ID: HGNC:28930 | Ensembl Gene: ENSG00000185640 | OMIM ID: 611160 |
| Drug and gene relationship at DGIdb | |||
Expression of KRT79:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KRT79 | 338785 | 1569909_at | 0.1221 | 0.6894 | |
| GSE26886 | KRT79 | 338785 | 1569909_at | 0.0383 | 0.7913 | |
| GSE45670 | KRT79 | 338785 | 1569909_at | -0.0458 | 0.6552 | |
| GSE53622 | KRT79 | 338785 | 78731 | -0.6360 | 0.0264 | |
| GSE53624 | KRT79 | 338785 | 78731 | -1.3335 | 0.0000 | |
| GSE63941 | KRT79 | 338785 | 1569909_at | 0.2056 | 0.1232 | |
| GSE77861 | KRT79 | 338785 | 1569909_at | -0.1649 | 0.1438 | |
| SRP007169 | KRT79 | 338785 | RNAseq | -4.3032 | 0.0000 | |
| SRP133303 | KRT79 | 338785 | RNAseq | -2.4546 | 0.0002 | |
| SRP159526 | KRT79 | 338785 | RNAseq | -2.6958 | 0.0032 | |
| TCGA | KRT79 | 338785 | RNAseq | 1.1011 | 0.1346 |
Upregulated datasets: 0; Downregulated datasets: 4.
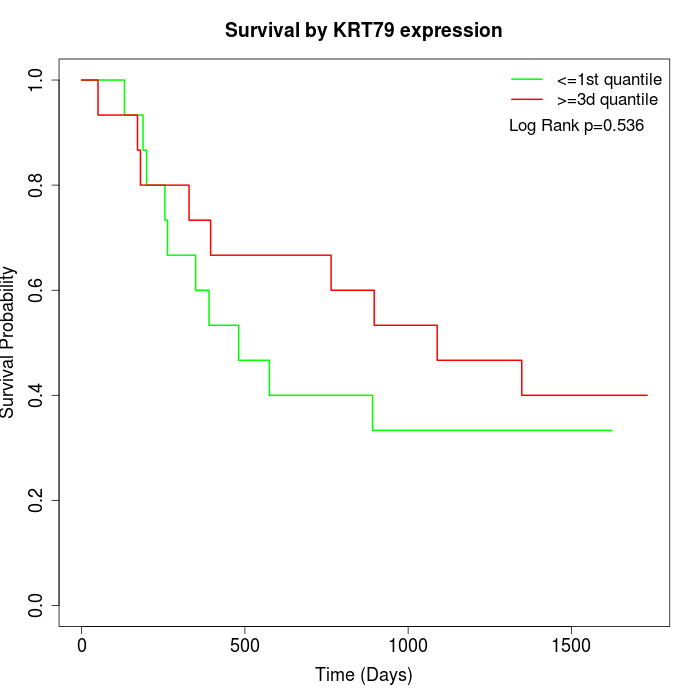
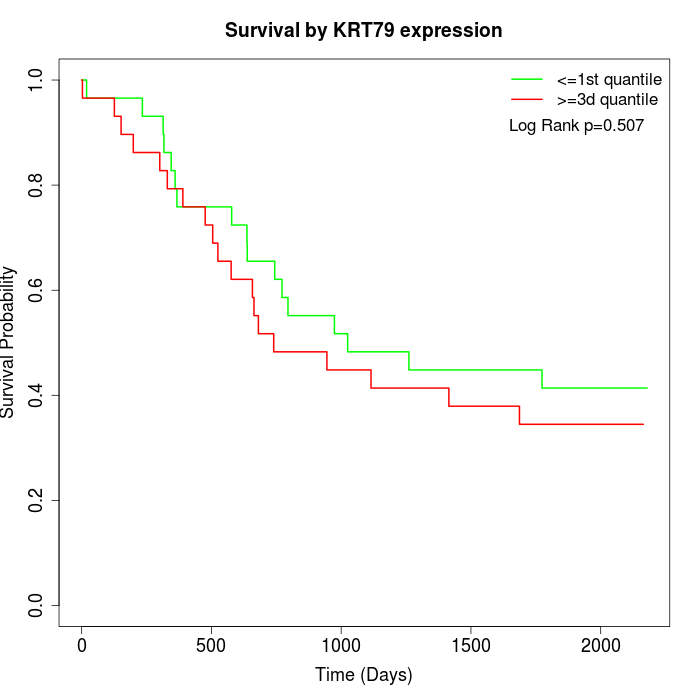
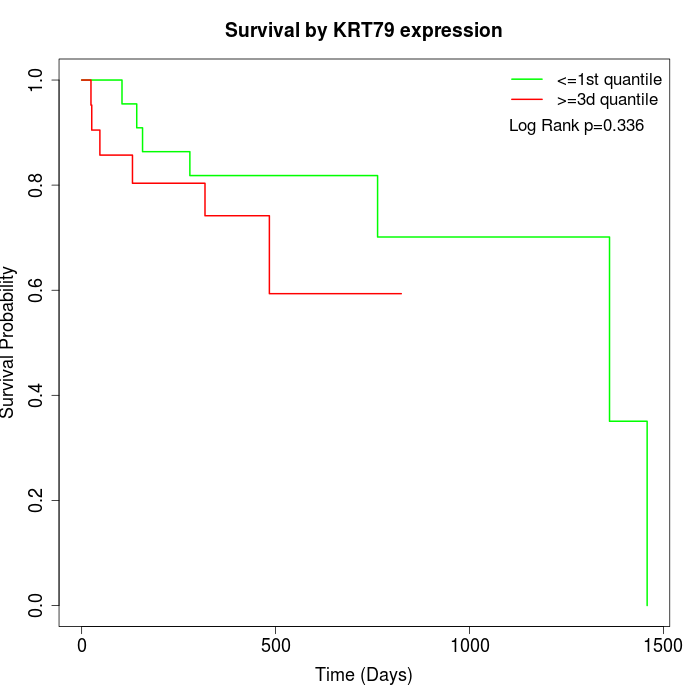
Survival by KRT79 expression:
Note: Click image to view full size file.
Copy number change of KRT79:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KRT79 | 338785 | 8 | 1 | 21 | |
| GSE20123 | KRT79 | 338785 | 8 | 1 | 21 | |
| GSE43470 | KRT79 | 338785 | 3 | 0 | 40 | |
| GSE46452 | KRT79 | 338785 | 7 | 1 | 51 | |
| GSE47630 | KRT79 | 338785 | 10 | 2 | 28 | |
| GSE54993 | KRT79 | 338785 | 0 | 5 | 65 | |
| GSE54994 | KRT79 | 338785 | 4 | 1 | 48 | |
| GSE60625 | KRT79 | 338785 | 0 | 0 | 11 | |
| GSE74703 | KRT79 | 338785 | 3 | 0 | 33 | |
| GSE74704 | KRT79 | 338785 | 5 | 1 | 14 | |
| TCGA | KRT79 | 338785 | 13 | 11 | 72 |
Total number of gains: 61; Total number of losses: 23; Total Number of normals: 404.
Somatic mutations of KRT79:
Generating mutation plots.
Highly correlated genes for KRT79:
Showing top 20/51 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KRT79 | ZNF214 | 0.699804 | 3 | 0 | 3 |
| KRT79 | ZGLP1 | 0.647766 | 3 | 0 | 3 |
| KRT79 | PSPN | 0.643923 | 3 | 0 | 3 |
| KRT79 | ZFHX2 | 0.640067 | 3 | 0 | 3 |
| KRT79 | C7orf61 | 0.631577 | 3 | 0 | 3 |
| KRT79 | VGLL2 | 0.628925 | 3 | 0 | 3 |
| KRT79 | IZUMO2 | 0.610954 | 3 | 0 | 3 |
| KRT79 | C11orf94 | 0.610098 | 3 | 0 | 3 |
| KRT79 | GPR26 | 0.609753 | 4 | 0 | 4 |
| KRT79 | HAO2 | 0.605521 | 3 | 0 | 3 |
| KRT79 | FGFR4 | 0.604496 | 4 | 0 | 3 |
| KRT79 | TMEM151B | 0.597284 | 3 | 0 | 3 |
| KRT79 | NOXA1 | 0.594506 | 3 | 0 | 3 |
| KRT79 | TUBGCP6 | 0.593115 | 3 | 0 | 3 |
| KRT79 | HUS1B | 0.59192 | 3 | 0 | 3 |
| KRT79 | TPSG1 | 0.590521 | 4 | 0 | 4 |
| KRT79 | MYLK2 | 0.588833 | 3 | 0 | 3 |
| KRT79 | CXCR5 | 0.581201 | 3 | 0 | 3 |
| KRT79 | ASPG | 0.578857 | 4 | 0 | 3 |
| KRT79 | EGFLAM-AS2 | 0.578202 | 3 | 0 | 3 |
For details and further investigation, click here