| Full name: C-X-C motif chemokine receptor 5 | Alias Symbol: MDR15|CD185 | ||
| Type: protein-coding gene | Cytoband: 11q23.3 | ||
| Entrez ID: 643 | HGNC ID: HGNC:1060 | Ensembl Gene: ENSG00000160683 | OMIM ID: 601613 |
| Drug and gene relationship at DGIdb | |||
CXCR5 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway |
Expression of CXCR5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CXCR5 | 643 | 216734_s_at | -0.1384 | 0.6895 | |
| GSE20347 | CXCR5 | 643 | 206126_at | -0.0788 | 0.2496 | |
| GSE23400 | CXCR5 | 643 | 206126_at | -0.0751 | 0.0300 | |
| GSE26886 | CXCR5 | 643 | 216734_s_at | -0.0522 | 0.7681 | |
| GSE29001 | CXCR5 | 643 | 206126_at | -0.0536 | 0.7685 | |
| GSE38129 | CXCR5 | 643 | 216734_s_at | -0.1544 | 0.0263 | |
| GSE45670 | CXCR5 | 643 | 216734_s_at | -0.0432 | 0.6748 | |
| GSE63941 | CXCR5 | 643 | 216734_s_at | -0.1175 | 0.4085 | |
| GSE77861 | CXCR5 | 643 | 216734_s_at | -0.0938 | 0.4861 | |
| GSE97050 | CXCR5 | 643 | A_24_P252945 | 0.1942 | 0.4116 | |
| TCGA | CXCR5 | 643 | RNAseq | -0.3698 | 0.3853 |
Upregulated datasets: 0; Downregulated datasets: 0.
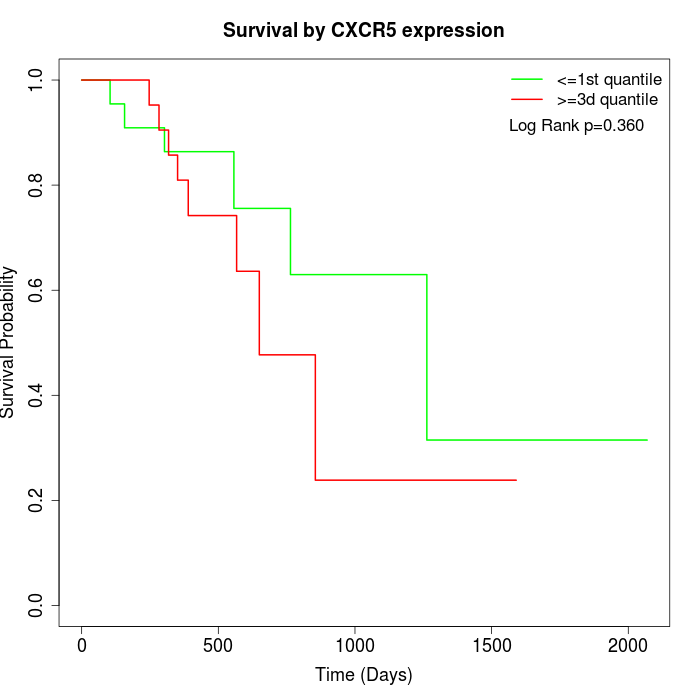
Survival by CXCR5 expression:
Note: Click image to view full size file.
Copy number change of CXCR5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CXCR5 | 643 | 2 | 11 | 17 | |
| GSE20123 | CXCR5 | 643 | 2 | 11 | 17 | |
| GSE43470 | CXCR5 | 643 | 2 | 8 | 33 | |
| GSE46452 | CXCR5 | 643 | 3 | 26 | 30 | |
| GSE47630 | CXCR5 | 643 | 2 | 19 | 19 | |
| GSE54993 | CXCR5 | 643 | 10 | 0 | 60 | |
| GSE54994 | CXCR5 | 643 | 5 | 19 | 29 | |
| GSE60625 | CXCR5 | 643 | 0 | 3 | 8 | |
| GSE74703 | CXCR5 | 643 | 1 | 5 | 30 | |
| GSE74704 | CXCR5 | 643 | 1 | 7 | 12 | |
| TCGA | CXCR5 | 643 | 4 | 51 | 41 |
Total number of gains: 32; Total number of losses: 160; Total Number of normals: 296.
Somatic mutations of CXCR5:
Generating mutation plots.
Highly correlated genes for CXCR5:
Showing top 20/733 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CXCR5 | NKX2-5 | 0.815005 | 3 | 0 | 3 |
| CXCR5 | ANKRD44 | 0.79669 | 3 | 0 | 3 |
| CXCR5 | FCHO1 | 0.786685 | 3 | 0 | 3 |
| CXCR5 | STK32B | 0.786677 | 3 | 0 | 3 |
| CXCR5 | TMEM214 | 0.783763 | 3 | 0 | 3 |
| CXCR5 | FBXO31 | 0.779958 | 3 | 0 | 3 |
| CXCR5 | RNF31 | 0.76887 | 3 | 0 | 3 |
| CXCR5 | GIT1 | 0.767922 | 3 | 0 | 3 |
| CXCR5 | TAGAP | 0.755286 | 3 | 0 | 3 |
| CXCR5 | BACH2 | 0.75187 | 3 | 0 | 3 |
| CXCR5 | PIM2 | 0.750072 | 3 | 0 | 3 |
| CXCR5 | NAGLU | 0.742956 | 4 | 0 | 4 |
| CXCR5 | ATP1A4 | 0.737098 | 3 | 0 | 3 |
| CXCR5 | MAST2 | 0.734511 | 3 | 0 | 3 |
| CXCR5 | HEYL | 0.733101 | 6 | 0 | 6 |
| CXCR5 | PZP | 0.728099 | 4 | 0 | 3 |
| CXCR5 | TCTN1 | 0.72435 | 3 | 0 | 3 |
| CXCR5 | AFF2 | 0.718543 | 3 | 0 | 3 |
| CXCR5 | ZFC3H1 | 0.718529 | 3 | 0 | 3 |
| CXCR5 | DNAJB12 | 0.718026 | 3 | 0 | 3 |
For details and further investigation, click here