| Full name: long intergenic non-protein coding RNA 687 | Alias Symbol: dJ1012F16.1 | ||
| Type: non-coding RNA | Cytoband: 20p12.2 | ||
| Entrez ID: 728450 | HGNC ID: HGNC:16194 | Ensembl Gene: ENSG00000228422 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LINC00687:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LINC00687 | 728450 | 232836_at | -0.0755 | 0.8149 | |
| GSE26886 | LINC00687 | 728450 | 232836_at | -0.1084 | 0.4337 | |
| GSE45670 | LINC00687 | 728450 | 232836_at | 0.1478 | 0.0693 | |
| GSE53622 | LINC00687 | 728450 | 19332 | 0.3064 | 0.0187 | |
| GSE53624 | LINC00687 | 728450 | 19332 | 0.1394 | 0.0827 | |
| GSE63941 | LINC00687 | 728450 | 232836_at | 0.1141 | 0.5273 | |
| GSE77861 | LINC00687 | 728450 | 232836_at | -0.1135 | 0.2408 |
Upregulated datasets: 0; Downregulated datasets: 0.
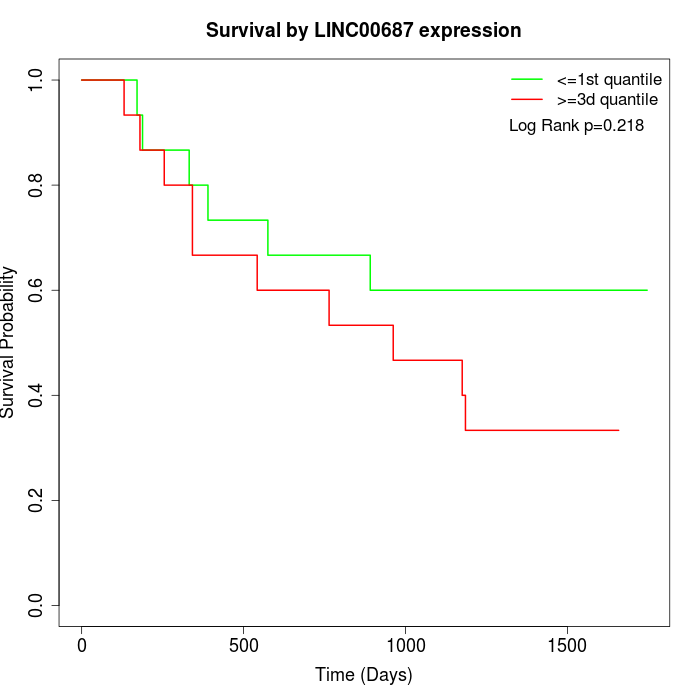
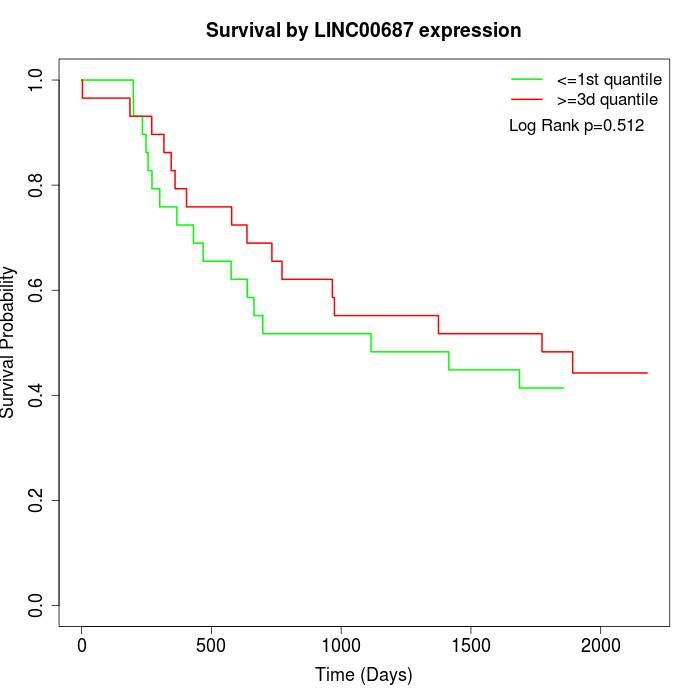
Survival by LINC00687 expression:
Note: Click image to view full size file.
Copy number change of LINC00687:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LINC00687 | 728450 | 9 | 1 | 20 | |
| GSE20123 | LINC00687 | 728450 | 9 | 1 | 20 | |
| GSE43470 | LINC00687 | 728450 | 11 | 1 | 31 | |
| GSE46452 | LINC00687 | 728450 | 26 | 1 | 32 | |
| GSE47630 | LINC00687 | 728450 | 18 | 4 | 18 | |
| GSE54993 | LINC00687 | 728450 | 1 | 14 | 55 | |
| GSE54994 | LINC00687 | 728450 | 25 | 1 | 27 | |
| GSE60625 | LINC00687 | 728450 | 0 | 0 | 11 | |
| GSE74703 | LINC00687 | 728450 | 8 | 1 | 27 | |
| GSE74704 | LINC00687 | 728450 | 5 | 1 | 14 | |
| TCGA | LINC00687 | 728450 | 43 | 10 | 43 |
Total number of gains: 155; Total number of losses: 35; Total Number of normals: 298.
Somatic mutations of LINC00687:
Generating mutation plots.
Highly correlated genes for LINC00687:
Showing top 20/142 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LINC00687 | FAM222A-AS1 | 0.738806 | 3 | 0 | 3 |
| LINC00687 | OPTC | 0.69926 | 4 | 0 | 4 |
| LINC00687 | GRIK4 | 0.677311 | 3 | 0 | 3 |
| LINC00687 | VGLL1 | 0.661671 | 3 | 0 | 3 |
| LINC00687 | OCM2 | 0.647927 | 3 | 0 | 3 |
| LINC00687 | LINC00293 | 0.647058 | 3 | 0 | 3 |
| LINC00687 | TXNDC2 | 0.640912 | 5 | 0 | 5 |
| LINC00687 | WDR87 | 0.630656 | 5 | 0 | 3 |
| LINC00687 | ZNF213-AS1 | 0.628649 | 3 | 0 | 3 |
| LINC00687 | TMEM235 | 0.625757 | 5 | 0 | 4 |
| LINC00687 | DSCAM-AS1 | 0.62407 | 5 | 0 | 4 |
| LINC00687 | C11orf72 | 0.622604 | 4 | 0 | 3 |
| LINC00687 | SPINT3 | 0.619555 | 4 | 0 | 3 |
| LINC00687 | PDZD7 | 0.618222 | 4 | 0 | 3 |
| LINC00687 | CDH24 | 0.616967 | 3 | 0 | 3 |
| LINC00687 | CHRNA2 | 0.609597 | 4 | 0 | 3 |
| LINC00687 | SLC22A24 | 0.604819 | 3 | 0 | 3 |
| LINC00687 | MYO18B | 0.603083 | 3 | 0 | 3 |
| LINC00687 | MC3R | 0.602434 | 3 | 0 | 3 |
| LINC00687 | CACNG4 | 0.600263 | 3 | 0 | 3 |
For details and further investigation, click here