| Full name: solute carrier family 22 member 24 | Alias Symbol: MGC34821|NET46 | ||
| Type: protein-coding gene | Cytoband: 11q12.3 | ||
| Entrez ID: 283238 | HGNC ID: HGNC:28542 | Ensembl Gene: ENSG00000197658 | OMIM ID: 611698 |
| Drug and gene relationship at DGIdb | |||
Expression of SLC22A24:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SLC22A24 | 283238 | 1553923_at | -0.0953 | 0.7871 | |
| GSE26886 | SLC22A24 | 283238 | 1553923_at | -0.1829 | 0.1788 | |
| GSE45670 | SLC22A24 | 283238 | 1553923_at | 0.0904 | 0.4402 | |
| GSE53622 | SLC22A24 | 283238 | 142951 | 0.1522 | 0.2229 | |
| GSE53624 | SLC22A24 | 283238 | 13534 | 0.3094 | 0.0017 | |
| GSE63941 | SLC22A24 | 283238 | 1553923_at | -0.2449 | 0.0582 | |
| GSE77861 | SLC22A24 | 283238 | 1553923_at | -0.0721 | 0.5467 |
Upregulated datasets: 0; Downregulated datasets: 0.
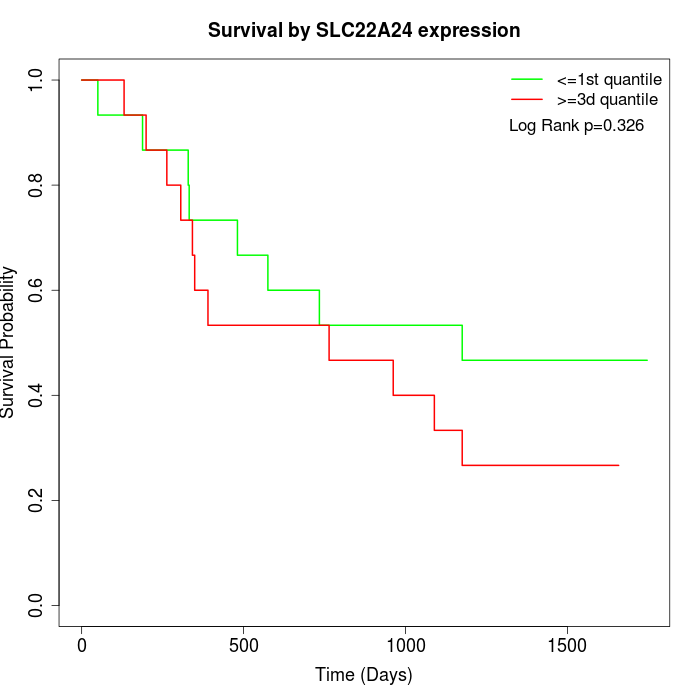
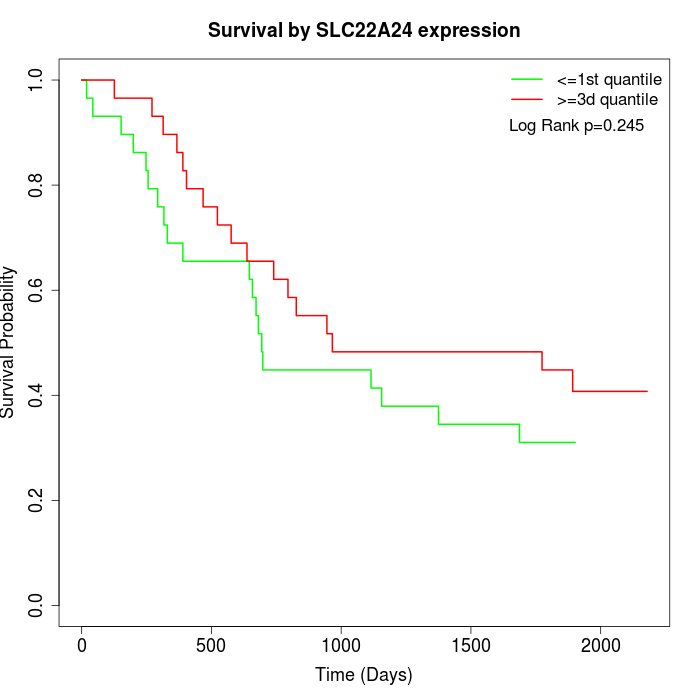
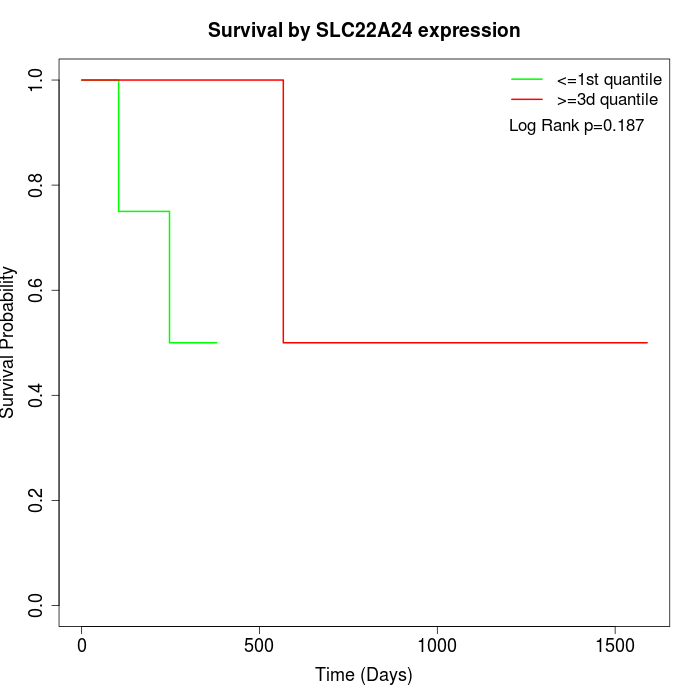
Survival by SLC22A24 expression:
Note: Click image to view full size file.
Copy number change of SLC22A24:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SLC22A24 | 283238 | 6 | 5 | 19 | |
| GSE20123 | SLC22A24 | 283238 | 7 | 4 | 19 | |
| GSE43470 | SLC22A24 | 283238 | 1 | 6 | 36 | |
| GSE46452 | SLC22A24 | 283238 | 8 | 4 | 47 | |
| GSE47630 | SLC22A24 | 283238 | 3 | 7 | 30 | |
| GSE54993 | SLC22A24 | 283238 | 3 | 0 | 67 | |
| GSE54994 | SLC22A24 | 283238 | 5 | 5 | 43 | |
| GSE60625 | SLC22A24 | 283238 | 0 | 3 | 8 | |
| GSE74703 | SLC22A24 | 283238 | 1 | 3 | 32 | |
| GSE74704 | SLC22A24 | 283238 | 5 | 3 | 12 | |
| TCGA | SLC22A24 | 283238 | 17 | 11 | 68 |
Total number of gains: 56; Total number of losses: 51; Total Number of normals: 381.
Somatic mutations of SLC22A24:
Generating mutation plots.
Highly correlated genes for SLC22A24:
Showing top 20/25 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SLC22A24 | SIX3 | 0.663068 | 3 | 0 | 3 |
| SLC22A24 | ACRBP | 0.620052 | 3 | 0 | 3 |
| SLC22A24 | LYPD8 | 0.616313 | 3 | 0 | 3 |
| SLC22A24 | LINC00687 | 0.604819 | 3 | 0 | 3 |
| SLC22A24 | LGALS13 | 0.595138 | 4 | 0 | 3 |
| SLC22A24 | SPAM1 | 0.58835 | 3 | 0 | 3 |
| SLC22A24 | CDH22 | 0.586971 | 5 | 0 | 3 |
| SLC22A24 | TMEM105 | 0.572778 | 5 | 0 | 3 |
| SLC22A24 | PHLPP2 | 0.572562 | 3 | 0 | 3 |
| SLC22A24 | TTTY11 | 0.558371 | 5 | 0 | 4 |
| SLC22A24 | SPHKAP | 0.557348 | 5 | 0 | 4 |
| SLC22A24 | MC3R | 0.555573 | 3 | 0 | 3 |
| SLC22A24 | CHRNA2 | 0.550626 | 5 | 0 | 4 |
| SLC22A24 | RPH3A | 0.547733 | 4 | 0 | 3 |
| SLC22A24 | LINC00421 | 0.540571 | 4 | 0 | 3 |
| SLC22A24 | CNTN5 | 0.540074 | 3 | 0 | 3 |
| SLC22A24 | CALML6 | 0.538097 | 4 | 0 | 3 |
| SLC22A24 | GLI4 | 0.536441 | 4 | 0 | 3 |
| SLC22A24 | CASP12 | 0.536036 | 4 | 0 | 3 |
| SLC22A24 | ARHGAP20 | 0.530977 | 3 | 0 | 3 |
For details and further investigation, click here